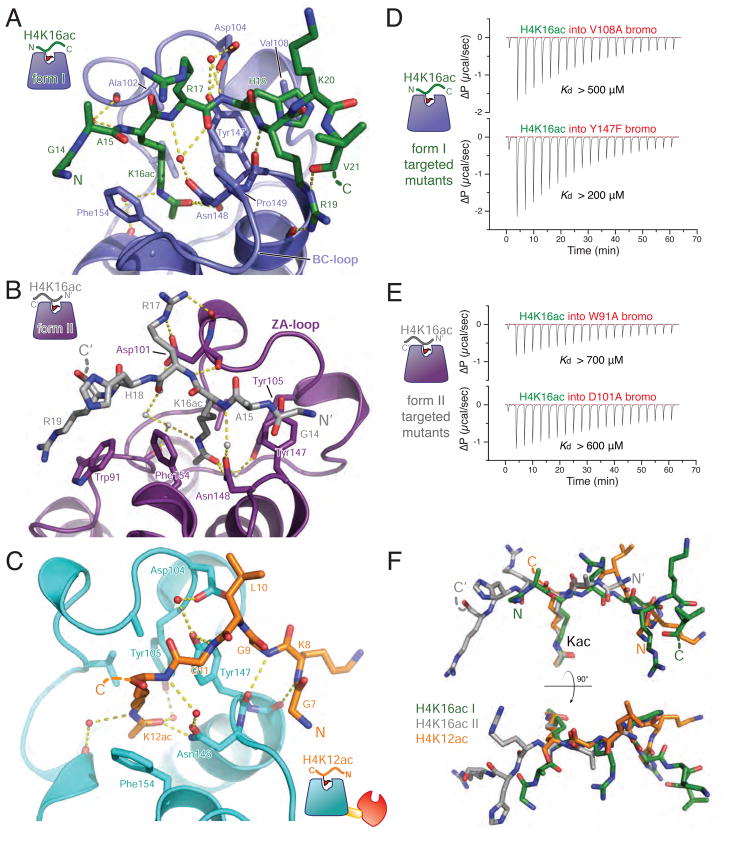
Figure 5. Structural analysis of the BPTF bromodomain peptide complexes.
(A) The model derived from crystal form I (bromodomain in blue) with the apical binding site for the H4K16ac peptide (green) rendered in ribbons and sticks. Hydrogen bonds are displayed as yellow dashed lines. (B) Interactions in crystal form II, the bromodomain is colored purple and the bound H4K16ac peptide is depicted in grey. (C) Interaction of the PHD-bromodomain in complex with H4K12ac peptide (peptide, orange; bromodomain, cyan). (D) ITC with BPTF bromodomain mutants designed to disrupt the form I binding mode (V108A and Y147F). Each of these mutations display modest binding deficits comporting with their modest roles in the form I interface. Dissociation constants were outside of the accurately measurable range, so a lower limit of possible Kd is provided for qualitative comparison. (E) Mutations to disrupt the form II binding interactions while leaving the form II binding mode intact: more substantial mutations, W91A and D101A, produce a more severe loss of affinity. For convenience of comparison, all ΔP scales are identical in scale. (F) Binding conformations of peptides (colored as in previous panels) from each of the structures is compared by Cα-superposition of their respective bromodomains. See also Figure S5.