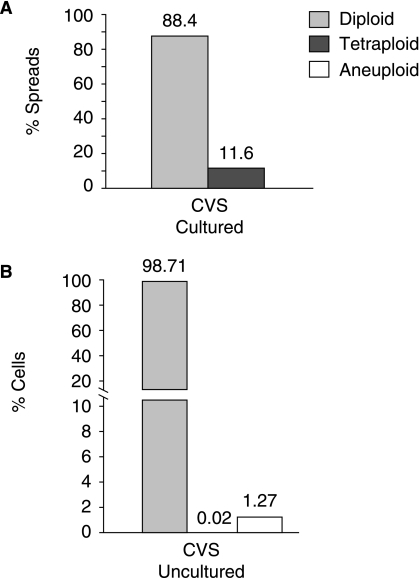
FIG. 4.
Chromosome stability in prenatal diagnostic samples. (A) Twenty three independent chorionic villus samples (C1–C23) were cultured for prenatal diagnosis. Chromosome spreads were prepared following 7–8 days (~PD 8–10) in culture to determine if numerical or structural abnormalities were detectable using the G-banding technique described in Fig. 1A. Twenty to thirty six chromosome spreads were analyzed in each CVS preparation (details are provided in Supplementary Table S3). Structural abnormalities were not detected (data not shown). The average percentages of diploid and tetraploid cells across all 23 CVS preparations are indicated. The tetraploid category includes spreads with close to double the normal chromosome number and may therefore encompass an aneuploid subset. (B) Uncultured cells from CVS preparations C1–C23 were dropped onto microscope slides. Each sample was hybridized with probes that detect chromosomes 13 and 21 (set 1) and also independently with a probe mix targeting chromosomes 18, X, and Y (set 2). A total of 200–400 interphase cells were scored in each sample. Half of the cells scored per sample were hybridized with each probe set and the frequency of diploid, tetraploid, and aneuploid cells recorded. A total of 4,900 cells were scored across the 23 CVS preparations. The average percentage of diploid, tetraploid, and aneuploid cells is presented. The average diploid rate was 98.71%. Tetraploidy was only observed in only one cell (in CVS sample C23 using probe set 1) giving a frequency of 0.02%. Cells in the aneuploid class averaged 1.27%.