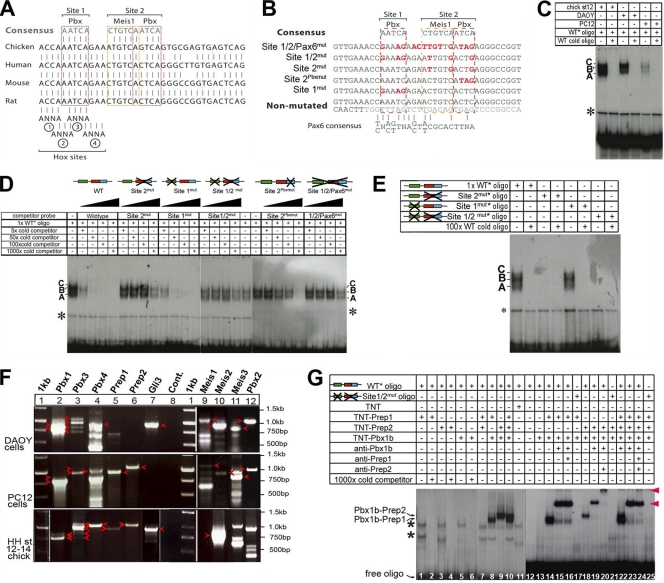
Fig. 3.
TALE proteins bind to HCNE1. (A) Sequence alignments of human, mouse, chicken, and rat genomic DNA show a high degree of conservation at sites 1 and 2, which contain binding sites for Pbx and Meis-Pbx, respectively. Four putative Hox binding site core sequences are also indicated, although no Pbx-Hox compound site was identified in databases. (B) Sequence of wild-type and mutated constructs and oligonucleotides used in EMSA and electroporation studies. Mutated nucleotides are indicated in red. (C) EMSA using a 35-bp oligonucleotide containing site 1 and site 2 incubated with nuclear extracts from chicken embryos and DAOY and PC12 cells in the presence/absence of a 1,000-fold excess of cold oligonucleotide. The asterisk indicates nonspecific binding. Complexes are labeled A, B, and C. (D) EMSA using a 35-bp oligonucleotide containing site 1 and site 2 incubated with nuclear extracts from chicken embryos in the presence of increasing amounts (5-, 50-, 100-, and 1,000-fold) of cold wild-type or mutated oligonucleotides. (E) EMSA using a 35-bp oligonucleotide containing no mutation or mutations in site 1, site 2, or sites 1 and 2 incubated with nuclear extracts from chicken embryos in the presence or absence of a 100-fold excess of cold wild-type oligonucleotide. (F) Expression of Pbx, Meis, and Prep genes was assessed using RT-PCR on mRNA extracted from DAOY cells, PC12 cells, and chicken embryos. Loading and expected sizes are as follows: lane 1, 1-kb DNA ladder; lane 2, 901 bp (Pbx1a) and 788 bp (Pbx1b); lane 3, 1,073 bp (Pbx3a), 960 bp (Pbx3b), 831 bp (Pbx3c), and 718 bp (Pbx3d); lane 4, 983 bp (Pbx4); lane 5, 959 bp (Prep1); lane 6, 1,059 bp (Prep2); lane 7, 929 bp (Gli3); lane 8, negative control; lane 9, 1,156 bp (Meis1a); lane 10, 934 bp (Meis2a); lane 11, 766 bp (Meis3); lane 12, 1,146 bp (Pbx2). Bands observed at the expected sizes and confirmed through sequencing are indicated with arrowheads. The other PCR products observed either are nonspecific bands or upon sequencing were found to be a paralogue (as is the case for the RT-PCR bands present in lane 12 that proved to not contain Pbx2 upon sequencing). Arrowheads indicate bands at the expected sizes. (G) EMSA using a 35-bp oligonucleotide containing either wild-type site 1 and site 2 or mutated site 1 and site 2 that was radiolabeled and incubated with in vitro-translated Pbx1b, Prep1, and Prep2 proteins in the presence or absence of anti-Pbx1b, anti-Prep1, or anti-Prep2 antisera. Asterisks indicate nonspecific binding bands.