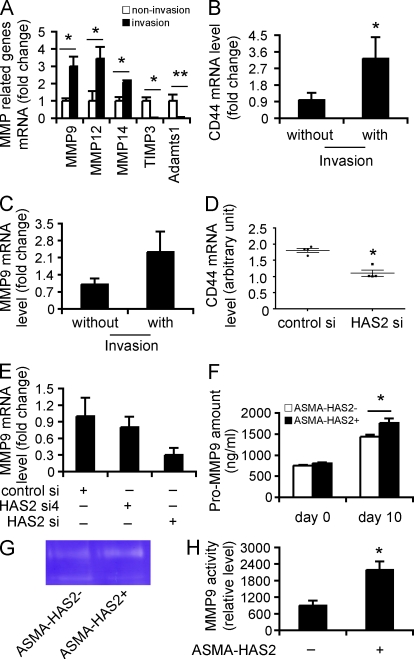
Figure 7.
HAS2 promotes fibroblast invasion by regulating CD44 and MMP expression and function. (A and B) RNA was extracted from invasive ASMA-HAS2+ fibroblasts, and 84 genes were analyzed by using a specialized qRT-PCR array for extracellular matrix synthesizing and degrading enzymes. RNAs from the fibroblasts that penetrated the filter in the absence of Matrigel were used as control. Representative genes up- or down-regulated in invasive ASMA-HAS2+ fibroblasts are shown (n = 5; *, P < 0.05; **, P < 0.01). (B) CD44 mRNA expression in invasive fibroblasts versus noninvasive fibroblasts from bleomycin-treated ASMA-HAS2+ lungs (n = 5; *, P = 0.05). (C) MMP9 mRNA expression in invasive IPF fibroblasts was compared with IPF fibroblasts that penetrated the filters in the absence of Matrigel by using real-time PCR (n = 5). The experiments were performed twice. (D) CD44 mRNA expression in HAS2 siRNA (HAS2 si)–transfected fibroblasts and control siRNA (control si) transfectants was determined using a microarray assay. The horizontal bars indicate the median expression values (n = 4; *, P < 0.05). (E) 48 h after HAS2 siRNA and control siRNA transfection, fibroblasts were cultured on Matrigel for an additional 6 h. mRNA was then extracted, and MMP9 mRNA expression was measured using real-time PCR. Data shown represent one of two separate experiments. (F) Fibroblasts from ASMA-HAS2− and ASMA-HAS2+ mice were cultured on Matrigel for 96 h, and pro-MMP9 protein in the media was measured using a pro-MMP9 ELISA kit (n = 3–7 per group; *, P < 0.05). The experiments were performed three times. (G) The media described in F was concentrated 10× using Microsep centrifugal devices. Equal amounts of protein were subjected to gelatin zymography for MMP9 activity. A representative image is shown. The experiments were repeated two times. (H) Quantification analysis of gelatin zymography for MMP9 activity results by using ImageJ software (n = 4–5 per group; *, P < 0.05). (A–F and H) Error bars indicate mean ± SEM.