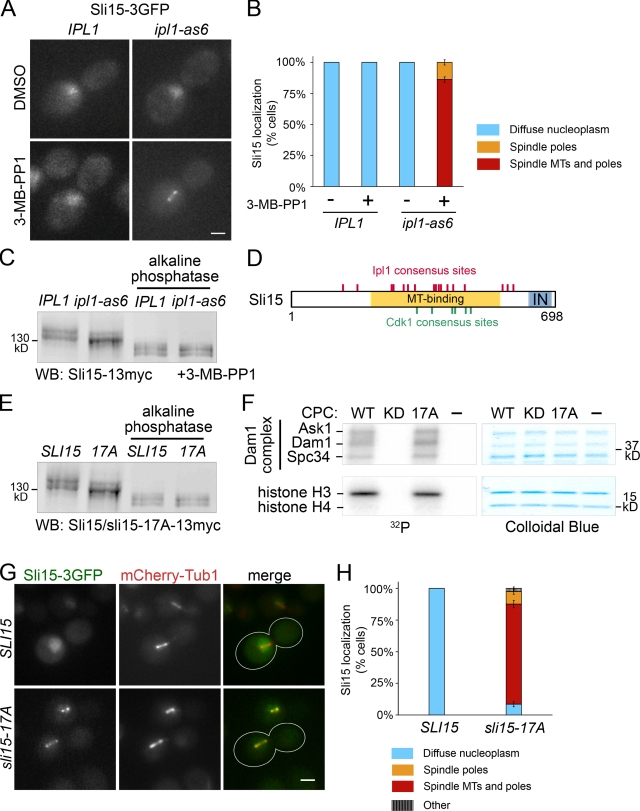
Figure 2.
Sli15/INCENP phosphorylation by Ipl1/Aurora is required for CPC exclusion from preanaphase spindles. (A) Ipl1 inhibition targets Sli15 to the preanaphase spindle. Representative cells containing IPL1 or ipl1-as6 and SLI15-3GFP, treated with DMSO or 40 µM 3-MB-PP1 for 15 min. Bar, 2 µm. (B) Quantification of observations represented in A. More than 200 cells from each treatment were analyzed using mCherry-Tub1 as a reference. Error bars show standard errors. (C) A subset of preanaphase Sli15 phosphorylation depends on Ipl1 activity in vivo. Sli15-13myc mobility shift upon Ipl1 inhibition was detected by Western blot of lysates from ipl1-as6 or IPL1 cells with or without alkaline phosphatase treatment. Cells were treated as described in A. (D) Diagram of Sli15 showing consensus sites for Ipl1/Aurora B ([RK]x[ST][ILVST]; Hsu et al., 2000; Bishop and Schumacher, 2002; Cheeseman et al., 2002) and Cdk1 ([ST]Px[RK]; Holmes and Solomon, 1996). MT-binding region (aa 227–559; Kang et al., 2001). T, three-helix bundle domain (aa 3–46; Jeyaprakash et al., 2007; Nakajima et al., 2009); IN, IN-box (aa 630–698; Adams et al., 2000). (E) Ipl1 phosphorylation site mutations in SLI15 diminish its phosphorylation in vivo. Western blot of whole-cell lysates from SLI15-13myc or sli15-17A-13myc cells. Cells were treated as described in A. (F) CPC containing sli15-17A retains normal kinase activity. In vitro kinase assay of the CPC containing only wild-type subunits (WT), ipl1-K133R (KD, kinase dead; Murata-Hori and Wang, 2002), or sli15-17A (17A) using the Dam1 complex and a histone H3/H4 tetramer as substrates. (G) sli15-17A shows a preanaphase spindle localization similar to that of Sli15 upon Ipl1 inhibition. Cells containing mCherry-TUB1, and SLI15-3GFP or sli15-17A-3GFP, were treated as described in A. Bar, 2 µm. (H) Quantification (as described in B) of the observations represented in G. “Other” denotes cells with foci that were not along spindle MTs or near the spindle poles. More than 200 cells from each treatment were analyzed. Error bars show standard errors.