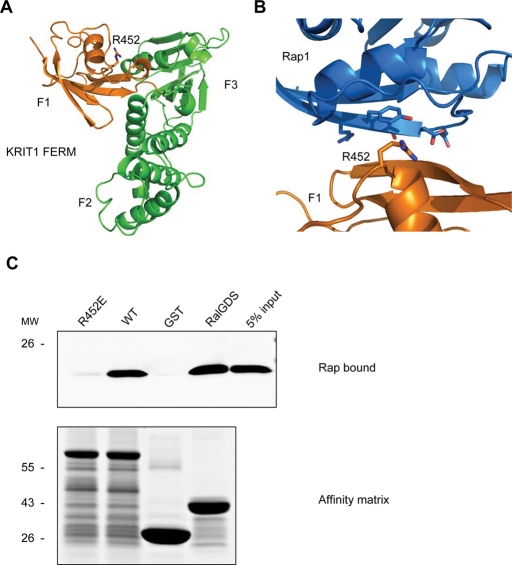
FIGURE 2:
KRIT1 FERM domain homologous modeling reveals R452 residue mediating KRIT1–Rap1 interaction. (A) Homology model of KRIT1 FERM domain. Residues 410–736 from KRIT1 were modeled using the moesin crystal structure as the template. The modeled structure is formed by three subdomains that correspond to the F1 (orange), F2, and F3 (both in green) subregions of the FERM fold. F1 subregion structurally resembles the c-Raf RBD in a complex with Rap1. An arginine residue (R-452), whose corresponding residue in c-Raf (R-89) mediates the c-Raf–Rap1 binding, is also conserved in the KRIT1 F1 subregion. (B) A closer look at the modeled binding interface between KRIT1 F1 subregion (orange) and Rap1 (blue). The R452 residue in KRIT1 and switch 1 domain residues in Rap1 are shown in black. (C) The R452 residue is crucial for KRIT1–Rap1 binding. GST-F123 WT pulls down recombinant Rap1V12 from HEK293 cell lysate (second lane). The GST-F123(R452E) mutant form, which changes a basic residue to an acidic residue, does not bind to Rap1V12 (left lane). Bottom, the equal loading of both GST fusion proteins as judged by SDS–PAGE and Coomassie Blue staining. Blots are representative of five experiments.