Figure 4.
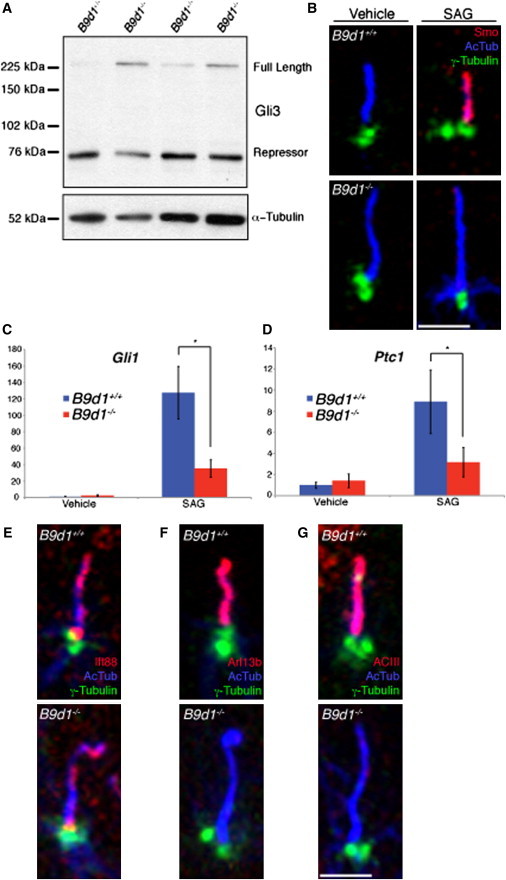
Hh Signaling and Protein Localization Defects in B9d1 Mutants
(A) Gli3 processing was assessed in E10.5 B9d1 heterozygote or mutant embryo lysates. Full-length Gli3 is stabilized in mutant lysates compared to the heterozygotes.
(B) B9d1 wild-type and mutant MEFs stained for cilia (AcTub, blue), basal bodies (γ-Tubulin, green), and Smo (red) in response to SAG treatment. Smo fails to localize to B9d1 mutant MEFs treated with SAG. Scale bar represents 2.5 μm.
(C and D) Gli1 and Ptc1 mRNA levels of B9d1 wild-type and mutant MEFs stimulated with SAG. The transcriptional response to SAG treatment is abrogated in B9d1 mutant MEFs. y axis values represent the fold change of mRNA compared to wild-type MEFs treated with vehicle only. Error bars represent the standard deviation. Asterisk indicates p < 0.0001.
(E–G) B9d1 wild-type and mutant MEFs stained for cilia (AcTub, blue), basal bodies (γ-Tubulin, green), and the ciliary proteins (red) Ift88 (E), Arl13b (F), and ACIII (G). Arl13b and ACIII fail to localize to B9d1 mutant cilia.
Scale bar represents 2.5 μm.
