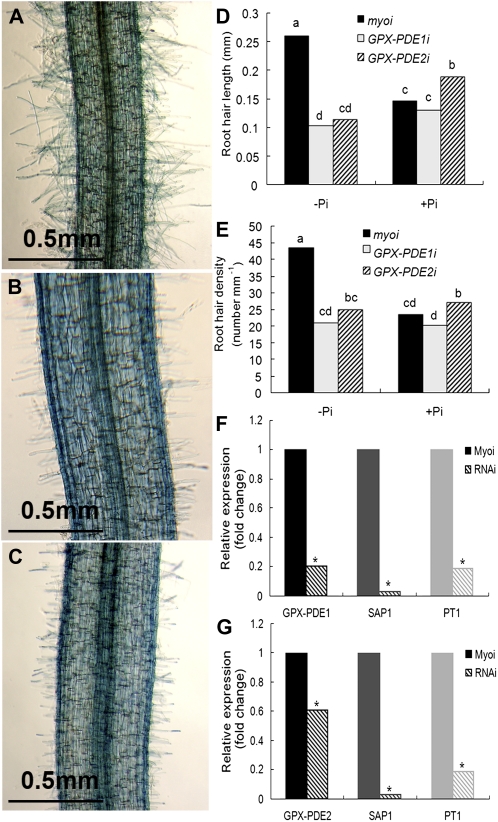
Figure 7.
Effect of GPX-PDE1 or GPX-PDE2 RNAi silencing on root hair development and expression of Pi starvation-induced genes. Reduced expression of GPX-PDE1 and GPX-PDE2 was achieved by subcloning a 631-bp mRNA sequence of GPX-PDE1 and a 510-bp mRNA sequence of GPX-PDE2 into an RNAi vector containing DsRED as a reporter and subsequent transformation to white lupin roots using A. rhizogenes. The human myosin gene was used as a control. Transgenic plants were grown under Pi-deficient conditions and harvested after 5 weeks. The DsRED reporter was used to screen transformed roots. A total of 48, 54, and 30 RNAi plants were evaluated for GPX-PDE1, GPX-PDE2, and myosin, respectively. A to C, Control RNAi (A), GPX-PDE1 RNAi (B), and GPX-PDE2 RNAi (C) showing root hair density and length from a zone 5 mature rootlet. D and E, Root hair length (D) and root hair density (E) of GPX-PDE1 and GPX-PDE2 RNAi zone 5 mature cluster rootlets show a significant reduction relative to the control myosin RNAi root hair length and density grown under Pi-deficient conditions. No significant differences were evident when grown under Pi-sufficient conditions. Lowercase letters denote significant differences between genes (P ≤ 0.05). Statistical significance was based on 11, 10, and eight biological replicates for myosin, GPX-PDE1, and GPX-PDE2 RNAi transgenic plants, respectively. F and G, qRT-PCR confirmed the reduction of the GPX-PDE1 (F) and GPX-PDE2 (G) transcript in the corresponding RNAi roots (patterned columns) relative to the control (solid columns). The transcript abundance of two other Pi deficiency-induced genes (SAP1 and PT1) was also reduced in GPX-PDE1 and GPX-PDE2 RNAi roots. Asterisks denote significant differences in expression levels relative to the control myosin RNAi gene (P ≤ 0.05).