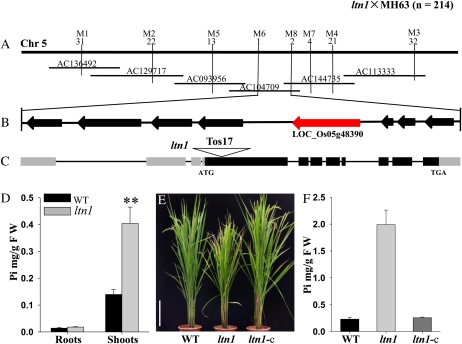
Figure 2.
Map-based cloning of the LTN1 gene. A, Preliminary and fine mapping. The LTN1 locus was preliminarily mapped to rice chromosome 5 (Chr 5) between markers M1 and M3. The gene was further delimited to a 54-kb genomic region between markers M6 and M8 within the bacterial artificial chromosome clone AC104709. The number of recombinants is marked corresponding to the molecular markers. B, The 54-kb interval has eight predicted ORFs, and the candidate gene is marked in red. C, LTN1 gene structure. Gray shading, untranslated region; black shading, ORF region; triangle, Tos17 transposon insertion. D, Pi concentration in 2-week-old seedlings of wild-type (WT; black bars) and ltn1 mutant (gray bars) plants grown under Pi-sufficient conditions (16 mg L−1). FW, Fresh weight. Error bars indicate sd (n = 5). Asterisks indicate the significance of differences between wild-type and ltn1 mutant plants as determined by Student’s t test: ** P < 0.01. E, Growth performance of wild-type, ltn1 mutant, and ltn1-complemented (ltn1-c) plants grown in the field for 3 months. Bar = 20 cm. F, Leaf Pi concentration in 3-month-old field-grown wild-type (black bar), ltn1 mutant (gray bar), and ltn1-complemented (dark gray bar) plants. Error bars indicate sd (n = 5).