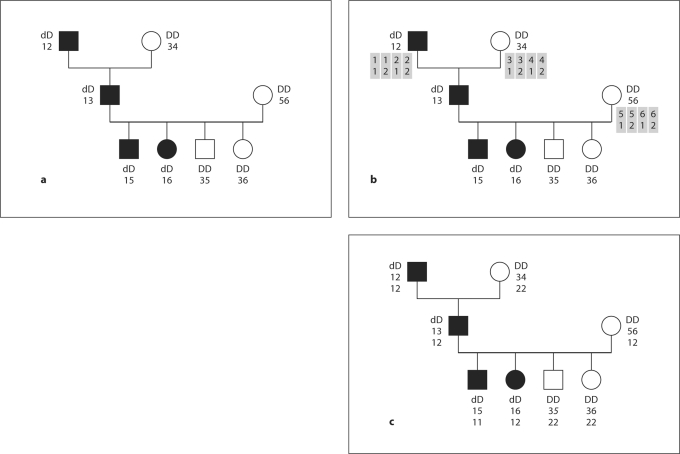
Fig. 1.
Three stages of sampling markers linked to a trait with SLINK and SUP. a The descent marker alleles (1, 2, 3, 4, 5, 6) are specified at the founders and the affection status may be specified in all individuals, if known. All alleles at the trait locus and the alleles of the non-founders at the descent marker locus are sampled by SLINK. b To begin filling in data at a SNP marker, SUP identifies possible haplotypes, which are shaded, combining the descent marker and a new SNP marker in the founders. In general if m − 1 markers are to be added, there would be 2m possible haplotypes. However, if some of the newly sampled markers are in LD with each other, then the probabilities of some haplotypes may be 0. c To complete the sampling, SUP does haplotype dropping from founders to non-founders, while modeling the recombination process. In this example, there is a single recombination between the descent marker and the new SNP marker, occurring in the maternal meiosis to the unaffected grandson, and those alleles are shown in italics.