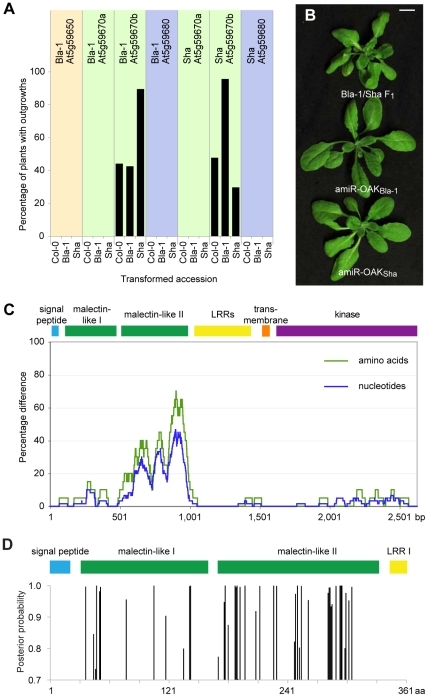
Figure 3. Identification of At5g59670b homologs as sufficient and necessary for outgrowths.
(a) Fraction of T1 plants (n≥90, except for Bla-1 transformed with Sha At5g59680 where n = 56) with outgrowths. (b) Suppression of outgrowths with amiRNAs against OAK (At5g59670b) from Bla-1 or Sha. (c) Divergence between OAK (At5g59670b) alleles from Bla-1 and Sha (sliding windows of 60 bp and 20 amino acids, respectively). (d) Identification of individual sites in the N-terminal part of OAK protein under positive selection (as determined by Bayesian Posterior Probability) across 34 accessions using PAML [43]. The second malectin-like domain is enriched for such sites.