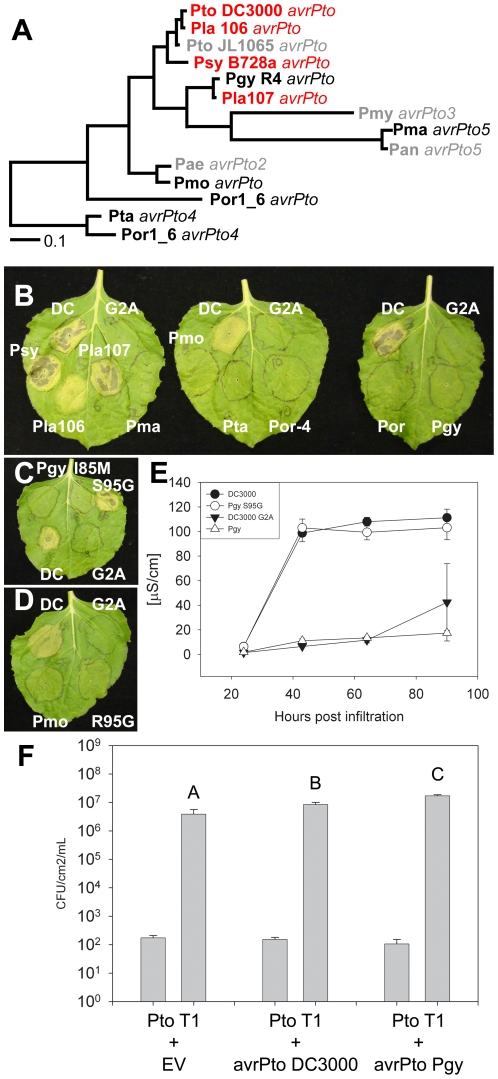
Figure 7. Phylogenetic analysis of the AvrPto superfamily reveals a residue required for avirulence function.
(A) Bayesian phylogeny for the AvrPto superfamily. Orthologs in red are recognized by Pto, orthologs in black are unrecognized, while orthologs in gray are untested. (B) Agrobacterium/N. benthamiana transient assay. Indicated AvrPto orthologs were co-expressed with Pto and symptoms are assessed at 4 days post innoculation. The Pto DC3000 AvrPto G2A mutant is a mislocalized negative control. (C) Mutation of S95G restores activity to the unrecognized ortholog AvrPtoPgy R4. Mutation of M85I does not result in recognition of AvrPtoPgy R4. (D) Mutation of R95G does not restore recognition of the AvrPtoPmo ortholog. (E) Ion leakage assay of Agrobacterium/N. Benthamiana transient inoculations. Error bars are one standard deviation. (F) Expression of either AvrPtoPto DC3000 or AvrPtoPgy R4 is sufficient to increase the virulence of Pto T1 on tomato plants incapable of recognizing AvrPto (tomato cultivar 76R prf3). Bars indicate growth at zero and four days after dip inoculation with 105 CFU/mL bacteria. Error bars are 2× standard error.