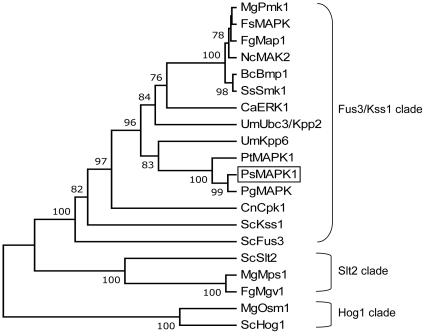
Figure 1. Phylogenetic analysis with PsMAPK1 and selected fungal MAP kinases (GenBank accession numbers in parenthesis).
Botrytis cinerea, BcBmp1 (AAG23132); Candida albicans, CaERK1 (P28869); Cryptococcus neoformans, CnCpk1 (Q8NK05); Fusarium solani, FsMAPK (AAB72017); Fusarium graminearum, FgMap1 (AAL73403) and FgMgv1 (AAM13670); Magnaporthe oryzae, MgPmk1 (AAC49521), MgMps1 (AAC63682) and MgOsm1 (AAF09475); Neurospora crassa, NcMAK2 (AAK25816); Puccinia striiformis f. sp. tritici, PsMAPK1 (HM535614); Puccinia triticina, PtMAPK1 (AAY89655); Puccinia graminis f. sp. tritici, PgMAPK (EFP88010); Saccharomyces cerevisiae, ScFus3 (CAA49292), ScHog1 (CAA97680), ScKss1 (CAA97038) and ScSlt2 (CAA41954); Sclerotinia sclerotiorum, SsSmk1 (AAQ54908); Ustilago maydis, UmKpp6 (CAD43731) and UmUbc3/Kpp2 (AAF09452). The unrooted phylogram was constructed based on NJ analysis. Confidence of groupings was estimated by using 1,000 bootstrap replicates. Numbers next to the branching point indicate the percentage of replicates supporting each branch.