Abstract
Forkhead Box M1 (Foxm1) is a transcription factor essential for organ morphogenesis and development of various cancers. Although complete deletion of Foxm1 in Foxm1−/− mice caused embryonic lethality due to severe abnormalities in multiple organ systems, requirements for Foxm1 in cardiomyocytes remain to be determined. This study was designed to elucidate the cardiomyocyte-autonomous role of Foxm1 signaling in heart development. We generated a new mouse model in which Foxm1 was specifically deleted from cardiomyocytes (Nkx2.5-Cre/Foxm1fl/f mice). Deletion of Foxm1 from cardiomyocytes was sufficient to disrupt heart morphogenesis and induce embryonic lethality in late gestation. Nkx2.5-Cre/Foxm1fl/fl hearts were dilated with thinning of the ventricular walls and interventricular septum, as well as disorganization of the myocardium which culminated in cardiac fibrosis and decreased capillary density. Cardiomyocyte proliferation was diminished in Nkx2.5-Cre/Foxm1fl/fl hearts owing to altered expression of multiple cell cycle regulatory genes, such as Cdc25B, Cyclin B1, Plk-1, nMyc and p21cip1. In addition, Foxm1 deficient hearts displayed reduced expression of CaMKIIδ, Hey2 and myocardin, which are critical mediators of cardiac function and myocardial growth. Our results indicate that Foxm1 expression in cardiomyocytes is critical for proper heart development and required for cardiomyocyte proliferation and myocardial growth.
Introduction
The heart is the first organ to function during embryonic development, the beating heart can be detected as early as embryonic day 8 (E8) in the mouse [1], [2]. Proper cardiac development requires strict adherence to a temporal and spatial pattern of gene expression. Embryonic development of the heart is mediated by proliferative growth, with cardiomyocytes rapidly progressing through the cell cycle and multiplying [3]. In the postnatal period, cardiomyocytes withdrawal from the cell cycle and cardiac growth becomes dependent on hypertrophy of individual cardiomyocytes [3]. Transcriptional regulation of cardiomyocyte proliferation during embryogenesis has been extensively studied, and several cardiac transcription factors were found to be critical for cardiomyocyte progression into the cell cycle. These include GATA family members 4 and 6 [4], myocardin [5], Twist family members 1 [6] and 2 [7], Hey2 [8], [9], Sox4 [10] and Nkx2.5 [11].
Foxm1 (previously known as HFH-11B, Trident, Win, or MPP2) is a member of the Forkhead Box (Fox) family of transcription factors which share homology in the Winged Helix/Forkhead DNA binding domain. Foxm1 is expressed in proliferating cells of all embryonic tissues, including cardiac progenitor cells and the early myocardium [12], [13]. However, expression wanes postnatally and Foxm1 can only be detected in a few adult tissues such as intestinal crypts, thymus and testis [14], [15]. Foxm1 signaling has been shown to be a critical mediator of both G1-S and G2-M transitions of the cell cycle, and to be upregulated in various human cancers [16], [17], [18], [19], [20], [21]. In addition, Foxm1 was determined to play a role in tissue repair following injury in the lungs and liver [15], [22], [23].
Foxm1-null (Foxm1−/−) mouse lines have been previously generated and characterized by two separate labs [24], [25]. Foxm1−/− mice in which the DNA binding and C-terminal transcriptional activation domains of the Foxm1 protein were deleted die in utero between E13.5 and E16.5 due to multiple abnormalities in various organ systems, including liver, lungs, blood vessels, brain and heart [13], [25], [26], [27]. Although these studies showed that Foxm1 plays a cell autonomous role for organ development in multiple cell types, the role of Foxm1 in cardiac development and function remains unknown. Given widespread organ defects in Foxm1−/− mice, it remains unclear whether Foxm1 is critical for heart development or if cardiac abnormalities are secondary to defects in other organ systems which could alter embryonic growth. Therefore, a direct role of Foxm1 in cardiomyocyte growth and/or function awaits elucidation.
As Foxm1 is widely expressed during embryogenesis [12], [28], [29], the recent focus has been to elucidate the cell-specific roles of Foxm1 in different tissues using conditional knockout mouse models. Specific deletion of Foxm1 from hepatoblasts resulted in embryonic lethality around day E18.5 with disruption of hepatic cords and vasculature, as well as a lack of intrahepatic bile ducts [25]. Deletion of Foxm1 from precursors of cerebellar granule neurons interfered with Shh-induced signaling to delay brain development [30]. Foxm1 deletion from T lymphocyte lineage decreased proliferation of early thymocytes and activated mature T cells without affecting apoptosis or T cell differentiation [31]. However, mice with endothelial- or macrophage-specific Foxm1 deletions developed normally [32], [33], indicating Foxm1 is dispensable in these cells lines during embryogenesis. Furthermore, while deletion of Foxm1 specifically from the pancreas did not affect pancreatic development [34], male mice developed islet dysfunction and diabetes resulting from impaired postnatal β-cell mass expansion [34] and females were prone to gestational diabetes [35], indicating Foxm1 requirements differ during pancreatic development. Deletion of Foxm1 specifically from smooth muscle cells did not affect differentiation, but mice died immediately after birth from severe pulmonary hemorrhage, structural defects in the arterial wall and esophageal abnormalities [28]. When Foxm1 was deleted conditionally in developing respiratory epithelium proliferation rates of respiratory epithelial cells were unaltered [12], suggesting that Foxm1 is not required for epithelial proliferation during lung development. However, deletion of Foxm1 from respiratory epithelium impaired lung maturation, decreased expression of surfactant-associated proteins SPA, SPB and SPC and delayed differentiation of type I cells from epithelial precursors causing respiratory failure after birth [12]. Thus, Foxm1 is essential for surfactant homeostasis and lung maturation during lung development. Studies in conditional Foxm1 knockout models have shown that Foxm1 plays unique roles in different tissues during embryonic development; the cardiomyocyte-specific role of Foxm1 in heart development remains unexplored.
In this study, we utilized the Cre-LoxP system to conditionally delete Foxm1 from cardiomyocytes to ascertain the cardiomyocyte-autonomous role of Foxm1 in heart development. Deletion of Foxm1 from cardiomyocytes resulted in chamber dilation and myocardial thinning, culminating in embryonic lethality in late gestation. Cardiac Foxm1 deletion caused decreased cardiomyocyte proliferation and altered expression of cell cycle regulators Cdc25B, Cyclin B1, nMyc, Plk-1 and p21cip1. We also identified CaMKIIδ, Hey2 and myocardin as new potential targets of Foxm1 signaling and mediators of myocardial thinning. This study shows that Foxm1 is critical for expression of cell cycle regulatory genes in developing cardiomyocytes and is required for proper heart development.
Methods
Foxm1 conditional knockout mice
We have previously described the generation of Foxm1 LoxP/LoxP (Foxm1fl/fl) mice, in which LoxP sequences flank exons 4 through 7 of the Foxm1 gene encoding the DNA binding and transcriptional activation domains [25]. Foxm1fl/fl mice were bred with Nkx2.5-Cre mice [36] to generate Nkx2.5-Cre/Foxm1fl/fl double transgenic mice with deletion of Foxm1 from the myocardium. Using lineage tracing experiments previous studies demonstrated that Nkx2.5-Cre was expressed in the early myocardium as well as epithelium of the first pharyngeal arch [36]. However, no gross morphological abnormalities were observed in any non-cardiac tissues examined including thyroid and thymus (data not shown). Nkx2.5-Cre/Foxm1fl/fl homozygous embryos exhibited an embryonic lethal phenotype with the exception of one mouse from an early breeding which survived to postnatal day 11 (P11). To generate Nkx2.5-Cre/Foxm1fl/fl embryos, Nkx2.5-Cre/Foxm1fl/+ heterozygous males were bred with Foxm1fl/fl females. Foxm1fl/fl or Nkx2.5-Cre/Foxm1fl/+ embryos from the same litter were used as controls. Animal studies were reviewed and approved by the Animal Care and Use Committee of Cincinnati Children's Hospital Research Foundation.
Immunohistochemical staining
Nkx2.5-Cre/Foxm1fl/fl and control embryos were harvested on embryonic day 14.5 (E14.5) and E17.5. Embryos were then fixed in 4% paraformaldehyde overnight and embedded into paraffin blocks. Paraffin sections of 5 µm were used for immunohistocheshow
Quantitative real-time RT-PCR (qRT-PCR)
Total cardiac RNA was prepared from individual Nkx2.5-Cre/Foxm1fl/fl and control hearts using RNA-STAT-60 (Tel-Test “B” Inc. Friendswood, TX). cDNA was generated using the Applied Biosystems High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA). Evaluation of expression levels of specific genes was performed by qRT-PCR using Taqman probes (Table 1) and StepOnePlus Real-Time PCR system (Applied Biosystems, Foster City, CA) as previously described [12], [20], [28].
Table 1. TaqMan gene expression assays (Applied Biosystems) used for qRT-PCR analysis.
| Mouse Foxm1 | Mm00514924_m1 |
| Mouse Cdc25B | Mm00499136_m1 |
| Mouse Cyclin B1 | Mm00838401_g1 |
| Mouse Cyclin D1 | Mm00432359_m1 |
| Mouse p21cip1 | Mm01303209_m1 |
| Mouse Plk-1 | Mm00440924_g1 |
| Mouse cMyc | Mm00487804_m1 |
| Mouse nMyc | Mm00476449_m1 |
| Mouse CaMKIIδ | Mm00499266_m1 |
| Mouse β-catenin | Mm00437992_m1 |
| Mouse NFATc3 | Mm01249200_m1 |
| Mouse GATA4 | Mm00484689_m1 |
| Mouse GATA6 | Mm00802636_m1 |
| Mouse Hey2 | Mm00469280_m1 |
| Mouse Myocardin | Mm00455051_m1 |
| Mouse Twist1 | Mm00442036_m1 |
| Mouse Twist2 | Mm00492147_m1 |
| Mouse Sox4 | Mm00486320_s1 |
| Mouse Notch1 | Mm00435245_m1 |
| Mouse Notch2 | Mm00440536_g1 |
| Mouse FGF10 | Mm00433275_m1 |
| Mouse IL-1β | Mm01336189_m1 |
| Mouse TGF-β1 | Mm03024053_m1 |
| Mouse BMP2 | Mm01340178_m1 |
| Mouse BMP4 | Mm01321704_m1 |
| Mouse Wnt5a | Mm00437347_m1 |
| Mouse Wnt7b | Mm00437357_m1 |
| Mouse β-actin | Mm00607939_s1 |
Western blot analysis
Hearts from E14.5 embryos were harvested and used to prepare protein extract. Three hearts from embryos with matching genotypes were pooled. Protein extracts were run on PAGE-SDS gels and transferred to PVDF membranes followed by incubation with primary antibodies specific for Foxm1 (1∶1000; C20; Santa Cruz), Cyclin B1 (1∶500; BD Pharmingen), or p21cip1 (1∶300; BD Pharmingen). Secondary antibodies were conjugated with horse-radish peroxidase. β-actin was used as a loading control.
Statistical analysis
Student's T-test was used to determine statistical significance. P values <0.05 were considered significant. Values for all measurements were expressed as mean±standard error of mean (SEM).
Results
Foxm1 protein and mRNA expression declines during embryonic and postnatal development
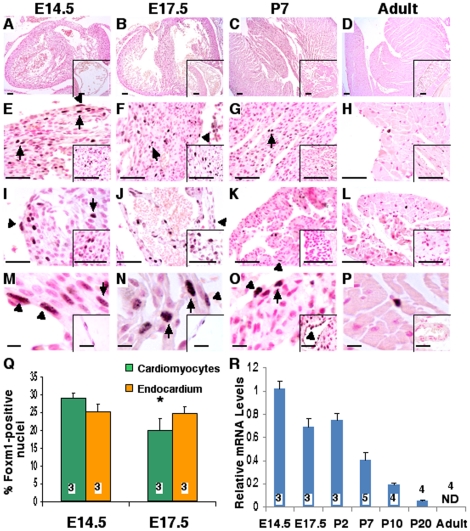
Hearts were collected from wild type mice at multiple timepoints during cardiac development. To identify cells expressing Foxm1, microtome sections of paraffin-embedded hearts were immunohistochemically stained using anti-Foxm1 antibodies. Foxm1 protein was highly abundant in the embryonic period and easily detectable in nuclei of cardiomyocytes and endothelial cells up to postnatal day 7 (P7) (Figure 1A–H). Foxm1 staining was observed throughout the developing heart, in the ventricles (Figure 1E–H and M–P), interventricular septum (insets of Figure 1E–H), atria (Figure 1I–L), heart valves (insets of Figure 1I–L), pericardium (insets of Figure 1M–N) and vasculature (insets of Figure 1O–P). The percentage of Foxm1-positive cardiomyocytes decreased from 29% at E14.5 to 20% at P7 (Figure 1Q) and nuclear Foxm1 staining was seldom observed in the adult heart as only 4±1% of cardiomyocytes were Foxm1-positive (data not shown). In addition, hearts were used to isolate total RNA and Foxm1 mRNA expression was examined by qRT-PCR. Similar to Foxm1 protein expression, Foxm1 mRNA was most highly expressed in the early embryonic period. Foxm1 expression declined by 30% from E14.5 to E17.5, but maintained a similar level of expression until P2. Foxm1 mRNA levels decreased rapidly in the postnatal period from P2 to P20, and Foxm1 mRNA was undetectable in the adult heart (11 weeks) (Figure 1R).
Figure 1. Foxm1 expression during heart development.
Foxm1 protein was evaluated in microtome sections of paraffin-embedded hearts from embryonic day 14.5 (E14.5) (A, E, I, M), E17.5 (B, F, J, N), postnatal day 7 (P7) (C, G, K, O), and adult wild type mice (D, H, L, P) by immunohistochemistry with Foxm1 antibodies [37] and counterstained with nuclear fast red (red nuclei). Foxm1 was detected in cardiomyocytes (arrows) and endocardial cells (arrowheads) from the ventricles (E–H, M–P), interventricular septum (inset E–H), and atrial walls (I–L) throughout cardiac development although expression progressively waned. Foxm1 was also detected in valves at E14.5 and E17.5 (inset I–J) but not postnatally (inset K–L). Foxm1-positive nuclei were detected in the embryonic pericardium (inset M–N) and in the coronary vasculature (inset O–P) until P7. The number of Foxm1-positive nuclei decreased during gestation in cardiomyocytes but was unaltered in endocardial cells (Q). Mean±SEM was determined from 5 random sections at E14.5 and P7 with 3 hearts in each group. qRT-PCR of total heart RNA demonstrated a decrease in Foxm1 mRNA from E14.5 to P20 and Foxm1 mRNA was undetectable in the adult heart (R). Foxm1 expression was normalized to β-actin mRNA. Significant differences (p<0.05) were indicated by asterisk. “N” values were represented by boxes inside or above bars. Scale bars represent 100 µm in A–D insets, 200 µm in A–D, 50 µm in E–L (main and insets) and 10 µm in M–P (main and insets).
Cardiomyocyte-specific deletion of Foxm1 causes embryonic lethality
Previous studies demonstrated that complete deletion of Foxm1 (Foxm1−/− mice) caused embryonic lethality between E13.5 and E16.5 and presented with malformation of multiple organ systems [25]. To ascertain the cardiomyocyte-autonomous role of Foxm1 signaling in cardiac development, Foxm1fl/fl mice (in which LoxP sites flank exons 4–7) were crossed with Nkx2.5-Cre mice to generate a mouse line in which the DNA binding and transcriptional activation domains of the Foxm1 protein were excised in cardiomyocytes (Figure 2A). Breeding pairs between Foxm1fl/fl and Nkx2.5-Cre/Foxm1fl/+ heterozygous mice were used to generate embryos with the Nkx2.5-Cre transgene and homozygous for the Foxm1fl/fl allele (Nkx2.5-Cre/Foxm1fl/fl) at an expected ratio of 1∶4. The majority of Nkx2.5-Cre/Foxm1fl/fl embryos did not survive to birth (Table 2). Although a near Mendelian ratio was observed for Nkx2.5-Cre/Foxm1fl/fl embryos at E14.5 (Table 2), between E14.5 and E17.5 nearly 50% of Nkx2.5-Cre/Foxm1fl/fl embryos were lost, with the remaining 50% lost between E17.5 and birth (Table 2). Only one Nkx2.5-Cre/Foxm1fl/fl pup survived to postnatal day 11 but exhibited severe growth retardation prior to harvest (data not shown). Thus, Foxm1 deletion from cardiomyocytes is sufficient to induce embryonic lethality.
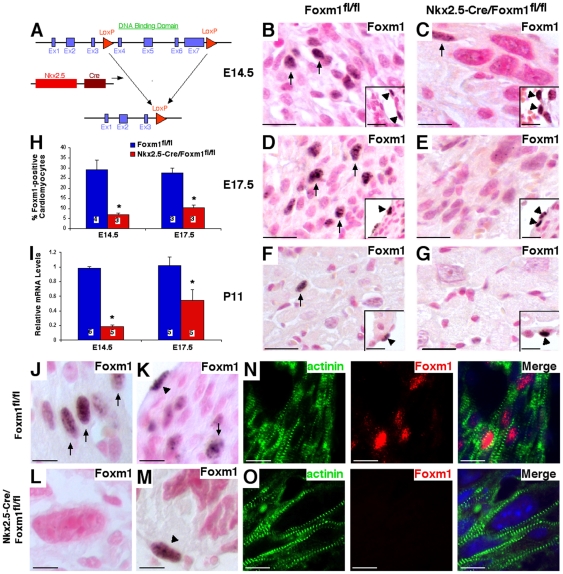
Figure 2. Generation of Nkx2.5-Cre/Foxm1fl/fl mice and efficiency of Foxm1 deletion.
The Cre-LoxP system was utilized to generate a mouse line in which Foxm1 was selectively deleted from myocardial cells (A). Foxm1fl/fl mice were crossed with mice expressing Nkx2.5-Cre to generate a mouse line in which Foxm1 is truncated in cardiomyocytes early in embryonic development. Microtome sections of paraffin-embedded hearts were prepared from Nkx2.5-Cre/Foxm1fl/fl and control (Foxm1fl/fl) mice at E14.5 (B–C, J–O), E17.5 (D–E) and P11 (F–G) and stained with Foxm1 antibodies. Foxm1 was observed in myocardial (arrows) and endocardial (arrowheads) cells. Quantification of the percentage of Foxm1-positive nuclei showed that cardiomyocytes positive for Foxm1 were significantly decreased in Nkx2.5-Cre/Foxm1fl/fl hearts compared to control at E14.5 and E17.5 (H). Similarly, Foxm1 mRNA was decreased in Nkx2.5-Cre/Foxm1fl/fl hearts at E14.5 and E17.5 as determined by qRT-PCR (I). Foxm1 expression was normalized to β-actin mRNA. Foxm1 was more readily detectable in cardiomyocytes from control (J–K, N) than Nkx2.5-Cre/Foxm1fl/fl hearts (L–M, O) as evidenced by colocalization of Foxm1-positive nuclei with α-actinin (N–O). However, Foxm1-positive endocardial cells could be detected in control (K) and Nkx2.5-Cre/Foxm1fl/fl hearts (M). Significant differences (p<0.05) were indicated by asterisk. “N” values were represented by boxes inside bars. Scale bars represent 200 µm in B–G and 50 µm in J–O.
Table 2. Breeding table Nkx2.5-Cre/Foxm1fl/+ X Foxm1fl/fl.
| E14.5 | E17.5 | Newborns | |
| Total embryos/pups | 31 | 46 | 67 |
| Expected ratio of Nkx2.5-Cre/Foxm1fl/fl | 0.25 | 0.25 | 0.25 |
| Experimental ratio of Nkx2.5-Cre/Foxm1fl/fl | 0.23 | 0.13 | 0.01 |
| % lethality of Nkx2.5-Cre/Foxm1fl/fl | 9.68 | 47.83 | 94.03 |
Cumulative breeding data between Nkx2.5-Cre/Foxm1fl/+ and Foxm1fl/fl mice shows significant lethality in Nkx2.5-Cre/Foxm1fl/fl embryos by day 17.5 (E17.5). 47% of Nkx2.5-Cre/Foxm1fl/fl embryos died by E17.5 and 94% lethality was observed in Nkx2.5-Cre/Foxm1fl/fl embryos at birth.
Efficiency of Foxm1 deletion from cardiomyocytes and endocardial cells was determined by immunohistochemical staining with antibodies against Foxm1. The percentage of Foxm1-positive cardiomyocyte nuclei was significantly decreased in Nkx2.5-Cre/Foxm1fl/fl embryos compared to littermate controls (Figure 2H). However, the percentage of Foxm1-positive endocardial cells was unchanged (41.2±2.7% vs. 40.9±5.4% at E14.5 and 37.1±1.4% vs. 38.8±1.3% at E17.5 for Foxm1fl/fl and Nkx2.5-Cre/Foxm1fl/fl respectively, Figure 2K and M, insets B–G), indicating specificity of Foxm1 deletion to cardiomyocytes. Cardiomyocyte Foxm1 staining was decreased by as much as 77% at E14.5 and was still decreased by nearly 63% at E17.5 (Figure 2H). The observed efficiency of Foxm1 deletion in Nkx2.5-Cre/Foxm1fl/fl hearts was substantiated by qRT-PCR analysis of total heart RNA. Compared to Foxm1fl/fl littermates, Foxm1 mRNA was decreased by 82% and 47% in Nkx2.5-Cre/Foxm1fl/fl hearts at E14.5 and E17.5, respectively (Figure 2I).
Embryonic deletion of Foxm1 from cardiomyocytes causes myocardial thinning, ventricular hypoplasia and disorganization of the myocardium
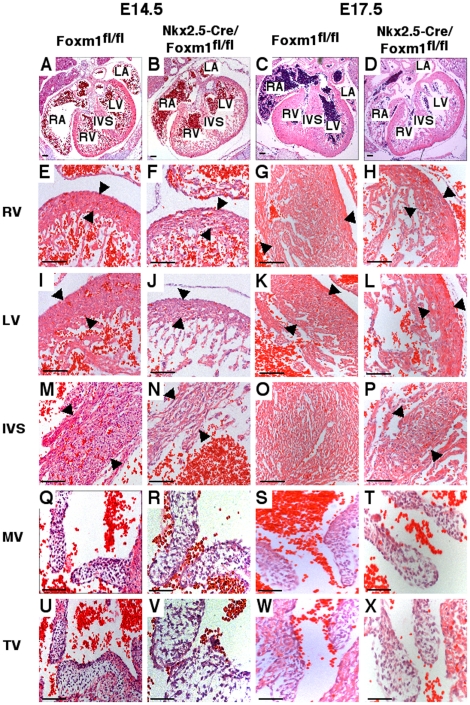
Hearts from Nkx2.5-Cre/Foxm1fl/fl embryos exhibited abnormal cardiac morphology in comparison to Foxm1fl/fl and Nkx2.5-Cre/Foxm1fl/+ littermate controls. The ventricular lumen was dilated and ventricular walls were thinner in Foxm1 deficient embryos compared to control embryos (Figure 3A–L). Ventricular wall thickness was significantly decreased at E14.5 and E17.5 (Figure 3E–L, Table 3). Thickness of the interventricular septum (IVS) was decreased by 42% and 62% at E14.5 and E17.5, respectively, (Figure 3M–P, Table 3) and cardiomyocyte organization within the IVS was in disarray (Figure 3M–P). There was, however, no significant change in the overall size of the heart at these two embryonic timepoints (Figure 3A–D). Furthermore, although an increased network of extracellular matrix was observed in valves from Nkx2.5-Cre/Foxm1fl/fl mice, there was no overall effect on valve size by deletion of Foxm1 from cardiomyocytes (Figure 3Q–X). In addition, there were no gross morphological changes in other organs known to delineate from Nkx2.5 expressing cells such as the thymus (data not shown).
Figure 3. Myocardial thinning and cardiomyocyte disarray in Nkx2.5-Cre/Foxm1fl/fl mice.
Microtome sections of paraffin-embedded hearts were prepared from Nkx2.5-Cre/Foxm1fl/fl and control Foxm1fl/fl mice at E14.5 and E17.5 and stained with hematoxylin and eosin (H&E). H&E staining showed that hearts from Nkx2.5-Cre/Foxm1fl/fl embryos possessed all four chambers and were similar in size to littermate control hearts (A–D). However, there was thinning of the muscular wall of the right ventricle (RV) (E–H), left ventricle (LV) (I–L), and interventricular septum (IVS) (M–P) in addition to disorganization of the musculature. There was no difference in the size of the leaflets in either the mitral (MV) (Q–T) or tricuspid valves (TV) (U–X). Arrowheads indicate area where measurements of thickness were made. Scale bars represent 100 µm in A–B and E–P, 200 µm in C–D and 50 µm in Q–X.
Table 3. Cardiac morphologic parameters.
| E14.5 | E17.5 | |||||
| RV (µm) | LV (µm) | IVS (µm) | RV (µm) | LV (µm) | IVS (µm) | |
| Foxm1fl/fl | 75.3±5.0 | 104.2±9.7 | 222.9±23.8 | 172.3±5.2 | 143.4±5.5 | 369.5±20.0 |
| Nkx2.5- Cre/Foxm1fl/fl | 63.0±8.4 | 59.6±5.1* | 142.2±24.2* | 98.5±4.4* | 94.6±7.4* | 159.1±12.6* |
Hearts from Nkx2.5-Cre/Foxm1fl/fl embryos had significantly decreased myocardial thickness compared to littermate control mice at E14.5 and E17.5. Five sections from 3 Foxm1fl/fl and Nkx2.5-Cre/Foxm1fl/fl embryos were measured to calculate mean±SEM. Significant differences (p<0.05) shown with asterisks. Abbreviations: RV, right ventricle; LV, left ventricle; IVS, interventricular septum.
Decreased cardiomyocyte proliferation in Nkx2.5-Cre/Foxm1fl/fl embryos
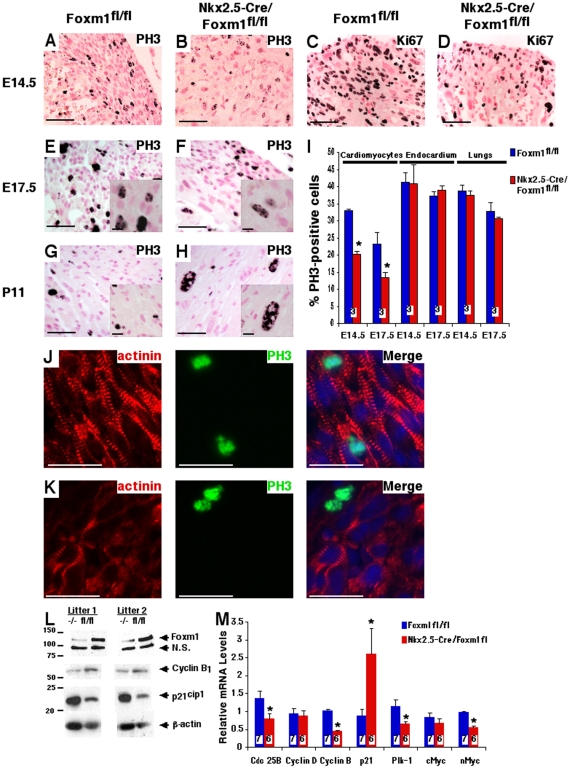
Cardiac proliferation was examined in Nkx2.5-Cre/Foxm1fl/fl and control embryos by immunohistochemical staining with antibodies against either phospho-histone 3 (PH3) or Ki-67. PH3-positive cardiomyocytes undergoing mitosis were observed in both Nkx2.5-Cre/Foxm1fl/fl and control Foxm1fl/fl mice at all timepoints studied (Figure 4A–H). However, cardiomyocyte proliferation was significantly diminished in Nkx2.5-Cre/Foxm1fl/fl hearts compared to littermate controls as indicated by significant decreases in the percentage of PH3-positive cardiomyocytes of 39% at E14.5 and 43% at E17.5 (Figure 4I). Yet, there was no change in cellular proliferation within the lung or in endocardial cells at these same timepoints (Figure 4I), indicating that proliferation defects were restricted to cardiomyocytes in Nkx2.5-Cre/Foxm1fl/fl embryos. Consistent with decreased numbers of PH3-positive cardiomyocytes, Nkx2.5-Cre/Foxm1fl/fl hearts displayed a dramatic reduction in Ki-67, a proliferation-specific protein (Figure 4C–D). Furthermore, decreased mRNA expression of cell cycle regulators Cdc25B, Cyclin B1, Polo-like kinase 1 (Plk-1) and nMyc was found in E14.5 Nkx2.5-Cre/Foxm1fl/fl hearts by qRT-PCR (Figure 4M). There was no change in the expression of Cyclin D1 or cMyc (Figure 4M). In addition, Nkx2.5-Cre/Foxm1fl/fl hearts displayed increased mRNA levels of p21cip1 (Figure 4M), a known cell cycle inhibitor critical for activity of cyclin-dependent kinase 2 (cdk2). Western blot analysis confirmed decreased protein levels of Foxm1 and Cyclin B1 as well as increased p21cip1 expression in Nkx2.5-Cre/Foxm1fl/fl hearts (Figure 4L). These results demonstrated that Foxm1 deletion from cardiomyocytes altered expression of cell cycle regulatory genes, contributing to proliferation defects and structural abnormalities in the developing heart.
Figure 4. Decreased cardiomyocyte proliferation in Nkx2.5-Cre/Foxm1fl/fl hearts.
Microtome sections of paraffin-embedded hearts were prepared from Nkx2.5-Cre/Foxm1fl/fl and control mice at E14.5 (A–D, J–K), E17.5 (E–F) and P11 (G–H) and stained for proliferation markers phospho-histone 3 (PH3) (A–B, E–H, J–K) and Ki-67 (C–D). The percent of PH3-positive cardiomyocytes was significantly lower in Nkx2.5-Cre/Foxm1fl/fl hearts compared to control at E14.5 and E17.5, though proliferation was unaltered in the lung and in endocardial cells (I). Proliferating cardiomyocytes were more prevalent in control (J) than in Nkx2.5-Cre/Foxm1fl/fl hearts (K) as evidenced by colocalization of PH3-positive nuclei with α-actinin. Consistent with decreased proliferation in Nkx2.5-Cre/Foxm1fl/fl hearts, there was decreased mRNA expression of the cell cycle regulators Cdc25B, Cyclin B1, Plk-1 and nMyc as determined by qRT-PCR (M). There was no change in the expression of Cyclin D1 or cMyc. Also consistent with decreased proliferation, there was increased mRNA expression of the cell cycle inhibitor p21cip1. Gene expression was determined using whole heart RNA and normalized to β-actin mRNA. Decreased gene expression of Foxm1, Cyclin B1, and increased p21cip1 expression translated to changes in protein levels as demonstrated by Western blot analysis (L). β-actin was used as a loading control. Significant differences (p<0.05) were indicated by asterisk. “N” values were represented by boxes inside bars. Scale bars represent 50 µm in A–H and J–K and 10 µm in insets E–H.
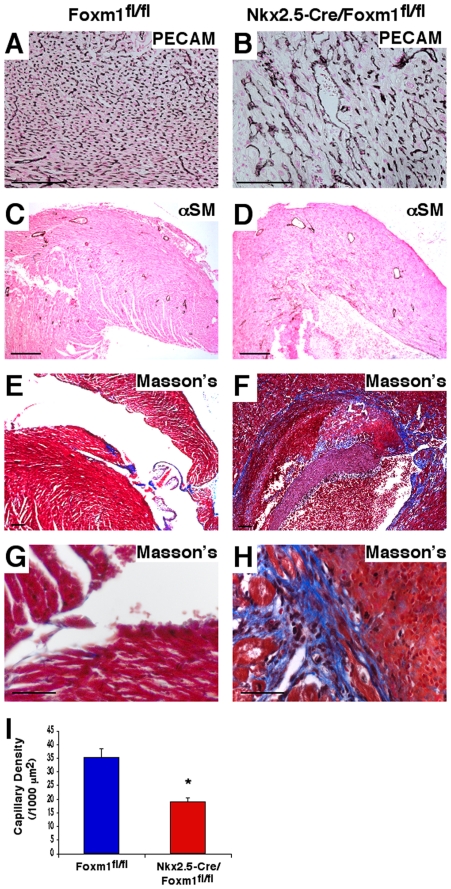
Decreased capillary density and cardiac fibrosis in postnatal Nkx2.5-Cre/Foxm1fl/fl hearts
Microtome sections of paraffin-embedded hearts from Nkx2.5-Cre/Foxm1fl/fl and control mice were subjected to immunohistochemical staining with antibodies against PECAM-1 or α-smooth muscle actin (αSM). PECAM-1 staining indicated a clear paucity in capillary density in the Nkx2.5-Cre/Foxm1fl/fl mouse heart at P11 (Figure 5A–B). Subsequent quantification showed a greater than 50% decrease in capillary density in the Nkx2.5-Cre/Foxm1fl/fl heart compared to Foxm1fl/fl control at P11 (Figure 5I). Despite the decreased capillary density, there was no difference in the number or morphology of coronary vessels in Nkx2.5-Cre/Foxm1fl/fl mouse hearts as indicated by αSM staining (Figure 5C–D). In addition, microtome sections from Nkx2.5-Cre/Foxm1fl/fl hearts were stained with Masson's Trichrome to detect cardiac fibrosis. Although there were no obvious signs of cardiac fibrosis at E17.5 (data not shown), the postnatal Nkx2.5-Cre/Foxm1fl/fl mouse heart displayed extensive fibrotic depositions in the interventricular septum as well as the apex of the ventricles (Figure 5F and H). No fibrosis was observed in postnatal Foxm1fl/fl control hearts (Figure 5E and G). Thus, Foxm1 deletion from cardiomyocytes altered vascular organization and induced cardiac fibrosis.
Figure 5. Decreased capillary density and cardiac fibrosis in Nkx2.5-Cre/Foxm1fl/fl heart.
Microtome sections of paraffin-embedded hearts were prepared from either Nkx2.5-Cre/Foxm1fl/fl or littermate control mice at P11 and stained for PECAM-1 (A–B), α-smooth muscle actin (αSM) (C–D) or Masson's Trichrome (E–H). Capillary density was decreased in Nkx2.5-Cre/Foxm1fl/fl hearts at P11 (A–B, I), while coronary vessel formation was not altered (C–D). Numbers of capillaries were counted in PECAM-stained hearts using 10 random sections and mean±SEM was determined to confirm decreased capillary density (I). Significant fibrosis was detected in the IVS and ventricular walls of Nkx2.5-Cre/Foxm1fl/fl hearts (F, H) while none was detected in control hearts (E, G). Significant differences (p<0.05) were indicated by asterisk. Scale bars represent 100 µm in A–F and 50 µm in G–H.
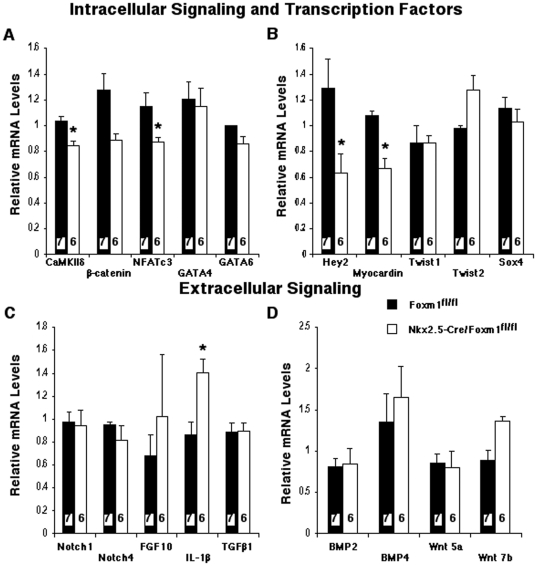
Foxm1 deletion from cardiomyocytes caused decreased expression of CaMKIIδ, NFATc3, myocardin and Hey2
To determine new potential Foxm1 target genes in the developing heart, expression of transcription factors and signaling molecules critical for heart morphogenesis or Foxm1 signaling was examined by qRT-PCR. Nkx2.5-Cre/Foxm1fl/fl embryos displayed a significant decrease in cardiac expression of NFATc3 (Figure 6A), a known Foxm1 target gene [13]. These results are consistent with reduced expression of NFATc3 in Foxm1-depleted ML-1 cells and hearts from Foxm1−/− mice [13]. Embryonic Nkx2.5-Cre/Foxm1fl/fl hearts exhibited normal mRNA expression of the intracellular signaling molecule β-catenin and the transcription factors GATA4, GATA6, Twist1, Twist2 and Sox4 (Figure 6A–B). Although interleukin-1β mRNA was increased in Nkx2.5-Cre/Foxm1fl/fl hearts (Figure 6C), there was no change in mRNA levels of other extracellular signaling molecules critical for heart morphogenesis, such as Notch1, Notch4, FGF10, TGF-β1, BMP2, BMP4, Wnt 5a or Wnt 7b (Figure 6C–D).
Figure 6. Reduced mRNA expression of CaMKIIδ, NFATc3, Hey2 and myocardin in Nkx2.5-Cre/Foxm1fl/fl hearts.
Whole-heart RNA was isolated from embryonic Nkx2.5-Cre/Foxm1fl/fl and control Foxm1fl/fl hearts and used for qRT-PCR analysis. Taqman primers specific for CaMKIIδ, β-catenin, NFATc3, GATA4, GATA 6, Hey2, myocardin, Twist1, Twist2, Sox4, Notch1, Notch4, FGF10, IL-1β, TGF-β1, BMP2, BMP4, Wnt5a and Wnt7b (Table 1) were used to evaluate cardiac mRNA expression. Gene expression was normalized to β-actin mRNA. Significantly diminished mRNA levels of the intracellular signaling molecule CaMKIIδ and transcription factors NFATc3, Hey2 and myocardin were detected in Nkx2.5-Cre/Foxm1fl/fl hearts. Increased mRNA expression of IL-1β was also detected. Significant differences (p<0.05) were indicated by asterisk. “N” values were represented by boxes inside bars.
Expression of calcium/calmodulin-dependent kinase IIδ (CaMKIIδ) was significantly reduced in Nkx2.5-Cre/Foxm1fl/fl hearts (Figure 6A). CaMKIIδ is an intracellular signaling molecule that is known to play a critical role in beat-to-beat cardiac physiology and has been shown to be an essential mediator of pressure overload and pharmacologically induced hypertrophy [37]. In addition, CaMKIIδ mediates calcium signaling and growth mechanisms within the cardiomyocyte [38]. Our results suggest that CaMKIIδ is a downstream effector of Foxm1 signaling as well as a contributing factor in myocardial thinning and embryonic lethality associated with cardiomyocyte-specific Foxm1 deletion.
Expression of both Hey2 and myocardin was reduced in Nkx2.5-Cre/Foxm1fl/fl hearts (Figure 6B). Hey2 knockout mice exhibited a thin walled left ventricle and decreased cardiomyocyte proliferation [8], similar to the phenotype in Nkx2.5-Cre/Foxm1fl/fl mice. Myocardin is a cofactor for the serum response factor (SRF) and is essential for cardiogenesis [39], [40]. Furthermore, SRF knockout mice exhibited a thin myocardium with dilated chambers and disorganized IVS [41], a phenotype similar to Nkx2.5-Cre/Foxm1fl/fl hearts. Therefore, decreased expression of Hey2 and myocardin may contribute to the cardiac abnormalities and embryonic lethality of the Nkx2.5-Cre/Foxm1fl/fl mouse. Altogether, our results demonstrate that Foxm1 deletion from cardiomyocytes alters expression of cardiac genes critical for heart morphogenesis and cardiomyocyte proliferation.
Discussion
We have previously generated and characterized a Foxm1-null (Foxm1−/−) mouse line in which mice die between embryonic days E13.5 and E16.5 due to severe defects in multiple organ systems including lungs, liver, heart and blood vessels [25]. We further demonstrated that tissue-specific deletion of Foxm1 from hepatocytes [25], respiratory epithelium [12] or smooth muscle cells [28] was sufficient to cause lethality in utero or shortly after birth. Therefore, Foxm1 is essential for organ morphogenesis in multiple organ systems. However, it remained to be determined whether cardiac malformation in Foxm1−/− embryos was due to cardiomyocyte derived effects of Foxm1 signaling or if these Foxm1−/− defects were indirect, resulting from abnormalities in other organ systems and altered embryonic homeostasis. To elucidate the cardiomyocyte-autonomous role of Foxm1 in embryonic heart development, we used the Cre-LoxP system to generate a conditional Foxm1 knockout mouse line in which Foxm1 is selectively deleted from cardiomyocytes under control of the Nkx2.5 promoter, one of the earliest known cardiac markers.
Deletion of Foxm1 from cardiomyocytes caused thinning and disorganization of the muscular walls of the heart, including both ventricles and the interventricular septum. Myocardial thinning was due to decreased cardiomyocyte proliferation accompanied by altered expression of multiple cell cycle regulatory genes. Ultimately, the myocardial hypoplasia in Nkx2.5-Cre/Foxm1fl/fl embryos caused lethality in late gestation. Although the myocardial phenotype exhibited similarities to that observed in hearts from Foxm1−/− mice, many unique features were observed in the conditional knockout model suggesting cardiomyocyte-autonomous roles for Foxm1 signaling. These include a delay in the onset of lethality, unaltered heart size, diminished cardiac capillary density and myocardial fibrosis. The decrease in cardiomyocyte proliferation in Nkx2.5-Cre/Foxm1fl/fl embryos was significantly less than in Foxm1−/− embryos suggesting that abnormalities in other cell types or tissues contributed to cardiac malformation in mice with complete deletion of Foxm1. Although we observed increased deposition of extracellular matrix in the atrio-ventricular valves of Nkx2.5-Cre/Foxm1fl/fl hearts the size of the valves was unaltered. These results are in contrast to valve thickening in Foxm1−/− mice [13] and suggest that although Foxm1 does mediate atrio-ventricular valve formation, this signaling is not cardiomyocyte-dependent. Alternatively, valve defects Foxm1−/− mice may result from altered blood pressure caused by structural abnormalities in blood vessels that were previously reported [26].
To date, embryonic lethality associated with ventricular hypoplasia and myocardial thinning has been linked to several signaling cascades including the transcription factor Hey2 [8], members of the NFAT family [42] or inactivation of serum response factor (SRF) [41]. In this study we described a model of myocardial thinning owing to multiple factors and resulting in embryonic lethality. In addition to altered expression of various cell cycle regulatory genes, this study identified Hey2, myocardin and CaMKIIδ as novel targets of Foxm1 signaling in vivo and as potential mediators of the thin ventricular phenotype.
We previously showed decreased expression of NFATc3 in Foxm1−/− hearts and in Foxm1-depleted cardiomyocytes in vitro [13]. This study confirmed that Foxm1 is a positive regulator of cardiac NFATc3 expression and further identified cardiomyocytes as the cell type responsible for Foxm1-regulated NFATc3 expression in vivo. It has been previously shown that dual deletion of NFATc3 and NFATc4 causes thin ventricles, decreased proliferation of ventricular myocytes and pericardial effusion culminating in embryonic lethality [42]. Therefore, decreased NFATc3 expression could be a contributing factor in myocardial thinning and embryonic lethality associated with Nkx2.5-Cre/Foxm1fl/fl mice.
Decreased expression of Hey2 can contribute to the cardiac phenotype and embryonic lethality of Nkx2.5-Cre/Foxm1fl/fl mice as evidenced by embryonic lethality and myocardial thinning in Hey2−/− mice, a phenotype similar to Nkx2.5-Cre/Foxm1fl/fl mice. Hey2 has also been shown to interact with the serum response factor (SRF) to inhibit activity of myocardin [43], which is essential for cardiogenesis, cardiomyocyte proliferation, migration and deposition of the extracellular matrix [39], [44], [45]. Furthermore, deletion of SRF from cardiomyocytes resulted in lethality between E10.5–13.5 with thin myocardium, dilated chambers and disorganized IVS [41], a phenotype similar to that observed in the Nkx2.5-Cre/Foxm1fl/fl hearts. Therefore, Foxm1 may directly influence myocardial development by decreasing expression of Hey2 and myocardin and possibly interfering with SRF-mediated signaling in cardiomyocytes.
CaMKIIδ deficiency caused augmented cardiac function in multiple heart injury models which manifested as severe alterations in cardiac structure. The finding of decreased CaMKIIδ mRNA in Nkx2.5-Cre/Foxm1fl/fl hearts suggests a role for Foxm1 in regulating cardiac CaMKIIδ expression. Interestingly, interleukin-1β (IL-1β) mRNA was increased in Nkx2.5-Cre/Foxm1fl/fl hearts. IL-1β has been reported to be a critical mediator of cardiac fibrosis [46]. Since significant fibrosis was observed in the postnatal Nkx2.5-Cre/Foxm1fl/fl heart, increased expression of IL-1β may contribute to fibrotic deposition.
In summary, we demonstrated that Foxm1 plays a cell-autonomous role in cardiomyocytes during cardiac development. Foxm1 deletion in developing cardiomyocytes caused embryonic lethality, decreased cardiomyocyte proliferation, diminished vascular density in the myocardium and induced cardiac fibrosis in the early postnatal period. This study further identified Hey2, myocardin, NFATc3, CaMKIIδ and various cell cycle regulatory genes as in vivo targets of Foxm1 signaling and potential mediators of the myocardial thinning and ventricular hypoplasia associated with the Nkx2.5-Cre/Foxm1fl/fl phenotype.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a grant from the National Institute of Health (HL 84151-04). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Horsthuis T, Christoffels VM, Anderson RH, Moorman AF. Can recent insights into cardiac development improve our understanding of congenitally malformed hearts? Clin Anat. 2009;22:4–20. doi: 10.1002/ca.20723. [DOI] [PubMed] [Google Scholar]
- 2.Savolainen SM, Foley JF, Elmore SA. Histology atlas of the developing mouse heart with emphasis on E11.5 to E18.5. Toxicol Pathol. 2009;37:395–414. doi: 10.1177/0192623309335060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Falco M, Cobellis G, De Luca A. Proliferation of cardiomyocytes: a question unresolved. Front Biosci (Elite Ed) 2009;1:528–536. doi: 10.2741/e49. [DOI] [PubMed] [Google Scholar]
- 4.Rojas A, Kong SW, Agarwal P, Gilliss B, Pu WT, et al. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol Cell Biol. 2008;28:5420–5431. doi: 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang RH, Zheng XL, Callis TE, Stansfield WE, He J, et al. Myocardin inhibits cellular proliferation by inhibiting NF-kappaB(p65)-dependent cell cycle progression. Proc Natl Acad Sci U S A. 2008;105:3362–3367. doi: 10.1073/pnas.0705842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vichalkovski A, Gresko E, Hess D, Restuccia DF, Hemmings BA. PKB/AKT phosphorylation of the transcription factor Twist-1 at Ser42 inhibits p53 activity in response to DNA damage. Oncogene. 2010;29:3554–3565. doi: 10.1038/onc.2010.115. [DOI] [PubMed] [Google Scholar]
- 7.Murakami M, Ohkuma M, Nakamura M. Molecular mechanism of transforming growth factor-beta-mediated inhibition of growth arrest and differentiation in a myoblast cell line. Dev Growth Differ. 2008;50:121–130. doi: 10.1111/j.1440-169X.2007.00982.x. [DOI] [PubMed] [Google Scholar]
- 8.Koibuchi N, Chin MT. CHF1/Hey2 plays a pivotal role in left ventricular maturation through suppression of ectopic atrial gene expression. Circ Res. 2007;100:850–855. doi: 10.1161/01.RES.0000261693.13269.bf. [DOI] [PubMed] [Google Scholar]
- 9.Xin M, Small EM, van Rooij E, Qi X, Richardson JA, et al. Essential roles of the bHLH transcription factor Hrt2 in repression of atrial gene expression and maintenance of postnatal cardiac function. Proc Natl Acad Sci U S A. 2007;104:7975–7980. doi: 10.1073/pnas.0702447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan X, Zhao J, Zhang WN, Li HY, Mu R, et al. Induction of SOX4 by DNA damage is critical for p53 stabilization and function. Proc Natl Acad Sci U S A. 2009;106:3788–3793. doi: 10.1073/pnas.0810147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeda M, Briggs LE, Wakimoto H, Marks MH, Warren SA, et al. Slow progressive conduction and contraction defects in loss of Nkx2-5 mice after cardiomyocyte terminal differentiation. Lab Invest. 2009;89:983–993. doi: 10.1038/labinvest.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalin TV, Wang IC, Meliton L, Zhang Y, Wert SE, et al. Forkhead Box m1 transcription factor is required for perinatal lung function. Proc Natl Acad Sci U S A. 2008;105:19330–19335. doi: 10.1073/pnas.0806748105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramakrishna S, Kim IM, Petrovic V, Malin D, Wang IC, et al. Myocardium defects and ventricular hypoplasia in mice homozygous null for the Forkhead Box M1 transcription factor. Dev Dyn. 2007;236:1000–1013. doi: 10.1002/dvdy.21113. [DOI] [PubMed] [Google Scholar]
- 14.Korver W, Roose J, Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997;25:1715–1719. doi: 10.1093/nar/25.9.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye H, Holterman AX, Yoo KW, Franks RR, Costa RH. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S phase. Mol Cell Biol. 1999;19:8570–8580. doi: 10.1128/mcb.19.12.8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalin TV, Wang IC, Ackerson TJ, Major ML, Detrisac CJ, et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66:1712–1720. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66:2153–2161. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida Y, Wang IC, Yoder HM, Davidson NO, Costa RH. The forkhead box M1 transcription factor contributes to the development and growth of mouse colorectal cancer. Gastroenterology. 2007;132:1420–1431. doi: 10.1053/j.gastro.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 20.Wang IC, Meliton L, Ren X, Zhang Y, Balli D, et al. Deletion of Forkhead Box M1 transcription factor from respiratory epithelial cells inhibits pulmonary tumorigenesis. PLoS One. 2009;4:e6609. doi: 10.1371/journal.pone.0006609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang IC, Meliton L, Tretiakova M, Costa RH, Kalinichenko VV, et al. Transgenic expression of the forkhead box M1 transcription factor induces formation of lung tumors. Oncogene. 2008;27:4137–4149. doi: 10.1038/onc.2008.60. [DOI] [PubMed] [Google Scholar]
- 22.Kalinichenko VV, Lim L, Shin B, Costa RH. Differential expression of forkhead box transcription factors following butylated hydroxytoluene lung injury. Am J Physiol Lung Cell Mol Physiol. 2001;280:L695–704. doi: 10.1152/ajplung.2001.280.4.L695. [DOI] [PubMed] [Google Scholar]
- 23.Kalinichenko VV, Gusarova GA, Tan Y, Wang IC, Major ML, et al. Ubiquitous expression of the forkhead box M1B transgene accelerates proliferation of distinct pulmonary cell types following lung injury. J Biol Chem. 2003;278:37888–37894. doi: 10.1074/jbc.M305555200. [DOI] [PubMed] [Google Scholar]
- 24.Korver W, Schilham MW, Moerer P, van den Hoff MJ, Dam K, et al. Uncoupling of S phase and mitosis in cardiomyocytes and hepatocytes lacking the winged-helix transcription factor Trident. Curr Biol. 1998;8:1327–1330. doi: 10.1016/s0960-9822(07)00563-5. [DOI] [PubMed] [Google Scholar]
- 25.Krupczak-Hollis K, Wang X, Kalinichenko VV, Gusarova GA, Wang IC, et al. The mouse Forkhead Box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesis. Dev Biol. 2004;276:74–88. doi: 10.1016/j.ydbio.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 26.Kim IM, Ramakrishna S, Gusarova GA, Yoder HM, Costa RH, et al. The forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J Biol Chem. 2005;280:22278–22286. doi: 10.1074/jbc.M500936200. [DOI] [PubMed] [Google Scholar]
- 27.Ueno H, Nakajo N, Watanabe M, Isoda M, Sagata N. FoxM1-driven cell division is required for neuronal differentiation in early Xenopus embryos. Development. 2008;135:2023–2030. doi: 10.1242/dev.019893. [DOI] [PubMed] [Google Scholar]
- 28.Ustiyan V, Wang IC, Ren X, Zhang Y, Snyder J, et al. Forkhead box M1 transcriptional factor is required for smooth muscle cells during embryonic development of blood vessels and esophagus. Dev Biol. 2009;336:266–279. doi: 10.1016/j.ydbio.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye H, Kelly TF, Samadani U, Lim L, Rubio S, et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17:1626–1641. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuller U, Zhao Q, Godinho SA, Heine VM, Medema RH, et al. Forkhead transcription factor FoxM1 regulates mitotic entry and prevents spindle defects in cerebellar granule neuron precursors. Mol Cell Biol. 2007;27:8259–8270. doi: 10.1128/MCB.00707-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue L, Chiang L, He B, Zhao YY, Winoto A. FoxM1, a forkhead transcription factor is a master cell cycle regulator for mouse mature T cells but not double positive thymocytes. PLoS One. 2010;5:e9229. doi: 10.1371/journal.pone.0009229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren X, Zhang Y, Snyder J, Cross ER, Shah TA, et al. Forkhead box M1 transcription factor is required for macrophage recruitment during liver repair. Mol Cell Biol. 2010;30:5381–5393. doi: 10.1128/MCB.00876-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao YY, Gao XP, Zhao YD, Mirza MK, Frey RS, et al. Endothelial cell-restricted disruption of FoxM1 impairs endothelial repair following LPS-induced vascular injury. J Clin Invest. 2006;116:2333–2343. doi: 10.1172/JCI27154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H, Ackermann AM, Gusarova GA, Lowe D, Feng X, et al. The FoxM1 transcription factor is required to maintain pancreatic beta-cell mass. Mol Endocrinol. 2006;20:1853–1866. doi: 10.1210/me.2006-0056. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Zhang J, Pope CF, Crawford LA, Vasavada RC, et al. Gestational diabetes mellitus resulting from impaired beta-cell compensation in the absence of FoxM1, a novel downstream effector of placental lactogen. Diabetes. 2010;59:143–152. doi: 10.2337/db09-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- 37.Zhang T, Miyamoto S, Brown JH. Cardiomyocyte calcium and calcium/calmodulin-dependent protein kinase II: friends or foes? Recent Prog Horm Res. 2004;59:141–168. doi: 10.1210/rp.59.1.141. [DOI] [PubMed] [Google Scholar]
- 38.Hempel P, Hoch B, Bartel S, Karczewski P. Hypertrophic phenotype of cardiac calcium/calmodulin-dependent protein kinase II is reversed by angiotensin converting enzyme inhibition. Basic Res Cardiol. 2002;97(Suppl 1):I96–101. doi: 10.1007/s003950200037. [DOI] [PubMed] [Google Scholar]
- 39.Chen JF, Wang S, Wu Q, Cao D, Nguyen T, et al. Myocardin marks the earliest cardiac gene expression and plays an important role in heart development. Anat Rec (Hoboken) 2008;291:1200–1211. doi: 10.1002/ar.20756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, et al. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–862. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 41.Parlakian A, Tuil D, Hamard G, Tavernier G, Hentzen D, et al. Targeted inactivation of serum response factor in the developing heart results in myocardial defects and embryonic lethality. Mol Cell Biol. 2004;24:5281–5289. doi: 10.1128/MCB.24.12.5281-5289.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bushdid PB, Osinska H, Waclaw RR, Molkentin JD, Yutzey KE. NFATc3 and NFATc4 are required for cardiac development and mitochondrial function. Circ Res. 2003;92:1305–1313. doi: 10.1161/01.RES.0000077045.84609.9F. [DOI] [PubMed] [Google Scholar]
- 43.Doi H, Iso T, Yamazaki M, Akiyama H, Kanai H, et al. HERP1 inhibits myocardin-induced vascular smooth muscle cell differentiation by interfering with SRF binding to CArG box. Arterioscler Thromb Vasc Biol. 2005;25:2328–2334. doi: 10.1161/01.ATV.0000185829.47163.32. [DOI] [PubMed] [Google Scholar]
- 44.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 45.Ueyama T, Kasahara H, Ishiwata T, Nie Q, Izumo S. Myocardin expression is regulated by Nkx2.5, and its function is required for cardiomyogenesis. Mol Cell Biol. 2003;23:9222–9232. doi: 10.1128/MCB.23.24.9222-9232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang MW, Matsumori A, Furukawa Y, Ono K, Okada M, et al. Neutralization of interleukin-1beta in the acute phase of myocardial infarction promotes the progression of left ventricular remodeling. J Am Coll Cardiol. 2001;38:1546–1553. doi: 10.1016/s0735-1097(01)01591-1. [DOI] [PubMed] [Google Scholar]