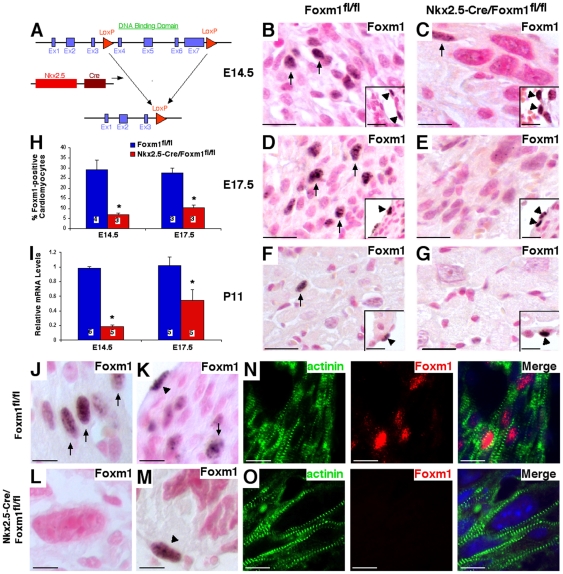
Figure 2. Generation of Nkx2.5-Cre/Foxm1fl/fl mice and efficiency of Foxm1 deletion.
The Cre-LoxP system was utilized to generate a mouse line in which Foxm1 was selectively deleted from myocardial cells (A). Foxm1fl/fl mice were crossed with mice expressing Nkx2.5-Cre to generate a mouse line in which Foxm1 is truncated in cardiomyocytes early in embryonic development. Microtome sections of paraffin-embedded hearts were prepared from Nkx2.5-Cre/Foxm1fl/fl and control (Foxm1fl/fl) mice at E14.5 (B–C, J–O), E17.5 (D–E) and P11 (F–G) and stained with Foxm1 antibodies. Foxm1 was observed in myocardial (arrows) and endocardial (arrowheads) cells. Quantification of the percentage of Foxm1-positive nuclei showed that cardiomyocytes positive for Foxm1 were significantly decreased in Nkx2.5-Cre/Foxm1fl/fl hearts compared to control at E14.5 and E17.5 (H). Similarly, Foxm1 mRNA was decreased in Nkx2.5-Cre/Foxm1fl/fl hearts at E14.5 and E17.5 as determined by qRT-PCR (I). Foxm1 expression was normalized to β-actin mRNA. Foxm1 was more readily detectable in cardiomyocytes from control (J–K, N) than Nkx2.5-Cre/Foxm1fl/fl hearts (L–M, O) as evidenced by colocalization of Foxm1-positive nuclei with α-actinin (N–O). However, Foxm1-positive endocardial cells could be detected in control (K) and Nkx2.5-Cre/Foxm1fl/fl hearts (M). Significant differences (p<0.05) were indicated by asterisk. “N” values were represented by boxes inside bars. Scale bars represent 200 µm in B–G and 50 µm in J–O.