Figure 1. Comparison of HPV DNA viral load and abundance of HPV-derived transcripts for established HPV-driven cancers versus normal skin and cuSCCs.
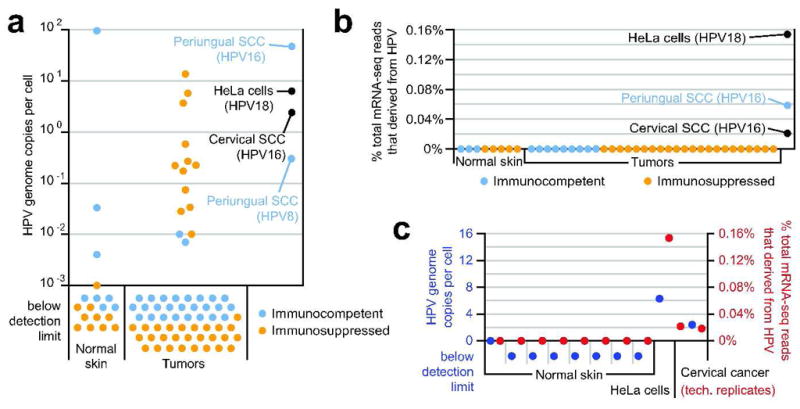
(A) HPV DNA viral loads determined by type-specific qPCR. For each sample with multiple type infection, the sum of the type-specific viral loads is shown (for details, see Table 2.) (B) Abundance of HPV-derived transcripts determined by mRNA-seq. Reads were filtered to remove host-derived and low-complexity sequence prior to viral mapping (see methods), and HPV counts are each presented as a percentage of their total dataset. The most frequently matched HPV type for the HeLa, periungual SCC, and cervical SCC samples is indicated. Cervical cancer is presented as the union of 2 technical replicate datasets. (C) Congruence between HPV genomic load (blue; presented as in panel A) and abundance of HPV-derived transcripts (red; presented as in panel B) among those control samples from which both types of data were collected.
