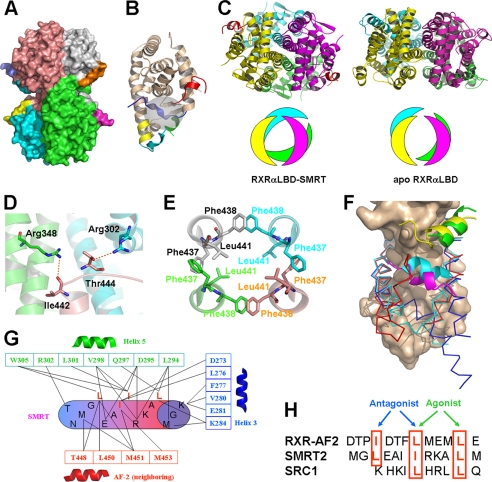
FIGURE 1.
Structure of RXRαLBD-SMRT. A, overall architecture of RXRαLBD-SMRT. Each RXRαLBD monomer and SMRT peptide is shown in a distinct color. B, RXRαLBD monomer indicating the key secondary structures for activation. wheat, conserved RXRαLBD core structure; green, N-terminal helix 3; yellow, helix 11; cyan, C-terminal AF-2 motif; blue, neighboring AF-2 motif; red, SMRT. Ligand-binding pockets are also indicated in the gray cycle. C, comparison of the tetramer rotations between the RXRαLBD-SMRT on the left and apoRXRαLBD on the right. Rotations are shown in both ribbon (upper) and model (lower) configurations. D, Ile442 and Thr444 forming two hydrogen bonds with Arg348 and Arg302 of the two neighboring monomers, respectively. E, Phe437 and Phe438 in the four monomers making hydrophobic interactions with each other in a hand-in-hand manner. Leu441 of each monomer extends itself inward to further stabilize helix 11 hydrophobically. F, structural superposition among apoRXRαLBD (blue), SMRT-bound RXRαLBD (red), and agonist-activated RXRαLBD (cyan) structures revealing different conformations of helix 3 and AF-2. The conserved core structure of the LBD is shown as surface; coactivator SRC-1 is in green, corepressor SMRT in yellow, and the AF-2 motif of the neighboring monomer in magenta. G, interactions between the SMRT motif (cylinder) and RXRαLBD (boxed residues, helix 3 in blue, helix 5 in green, and the AF-2 motif of the neighboring monomer in red). H, structure-based sequence alignment of the AF-2 motif, corepressor SMRT, and coactivator SRC1 on the overlapped binding pocket. The key residues are shown in red boxes and are indicated for the agonist or antagonist binding.