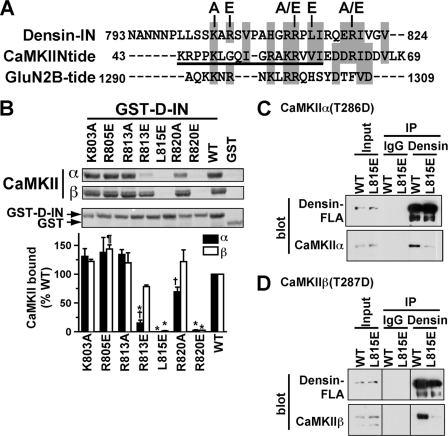
FIGURE 6.
L815E mutation of the densin IN domain disrupts CaMKII binding. A, amino acid sequences of the densin-IN domain, the CaMKIIN inhibitory domain, and the CaMKII binding domain of GluN2B are aligned with identical or similar residues in gray boxes. The underlined region of CaMKIIN-tide was resolved in an x-ray crystal structure of the CaMKII-CaMKIIN-tide complex (44). Densin-IN domain mutants characterized in panel B are indicated above. A color version of this panel is available online as supplemental Fig. 1. B, Thr-286/287-autophosphorylated CaMKIIα or CaMKIIβ were incubated with GST-D-IN (WT or mutated) or GST. Complexes isolated on glutathione-agarose were analyzed by protein staining of nitrocellulose membranes. Binding of CaMKII isoforms to mutated GST-D-INs was expressed as a percentage of the binding to WT (mean ± S.E., n = 3). †, p < 0.05 compared with binding of CaMKIIβ (2-way analysis of variance with Bonferroni post-test). *, p < 0.001 compared with WT (1-way analysis of variance with Dunnett's multiple comparison test). ¶, p < 0.05 compared with WT (1-way analysis of variance with Dunnett's multiple comparison test). C, extracts of HEK293 cells expressing mCherry-CaMKIIα-T286D and GFP-tagged densin-FLA (WT or L815E) were incubated with densin Ab450 or control goat IgG. Inputs and immune complexes were immunoblotted for densin and CaMKII. IP, immunoprecipitated. D, lysates from HEK293 cells co-expressing mCherry-CaMKIIβ-T287D with GFP-tagged densin-FLA (WT or L815E) were analyzed as in panel C. Samples were analyzed in parallel with those in panel C but were loaded in a different order to avoid confusion during analysis. The Input, IgG, and Densin lane pairs were each flipped horizontally during preparation of the figure so that all lanes are in the same order as in panel C; lines on the blots indicate these discontinuities.