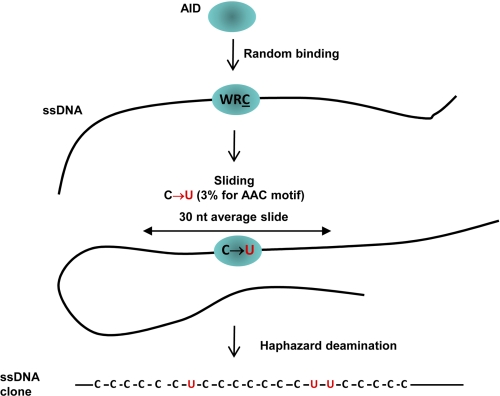
FIGURE 6.
Random walk model depicting AID scanning and catalyzing haphazard and inefficient deamination of C to U on ssDNA. AID binds at a random location on ssDNA. AID scans in accord with a modified random walk model (see text) in which the length of a slide in either direction is determined by a geometric distribution, i.e. shorter slides are favored over longer slides. An excellent fit of the model to the data shows that AID only rarely deaminates C to U, at about 3% efficiency for even “preferred” AAC hot motifs, thus ensuring mutational diversity. Deaminations occur predominantly during sliding. Intramolecular jumping/hopping (Fig. 3d) or inter-segmental transfer can also occur to reposition AID to another location on the same ssDNA molecule without altering the deamination patterns (see text). Deaminations occur mostly as singletons, although deamination clusters arise less frequently during repeated sliding over the same region of ssDNA. The average length of a random slide is 30 nt (10 trinucleotide motifs).