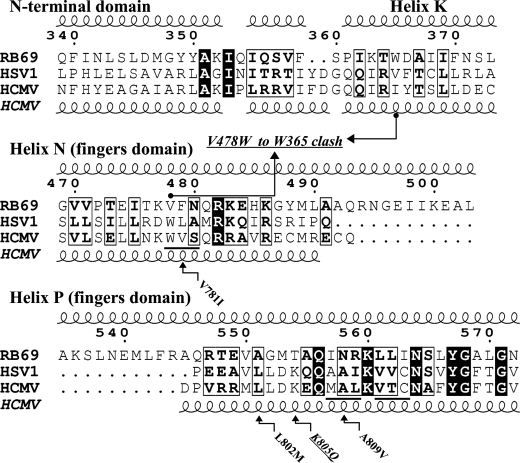
FIGURE 4.
Annotated sequence alignment (50) of family B polymerases from bacteriophage RB69, HSV1, and HCMV. Residue numbers for RB69 are shown. The structure-based sequence alignment between RB69 and HSV1 is based on their apo structures (Protein Data Bank codes 1IH7 and 2GV9) (15, 29). Secondary structure elements depicted at the top correspond to RB69 gp43, whereas helices shown at the bottom are predicted for HCMV UL54 from the HSV1 structure. The three HCMV blocks underlined comprise the nine chimeric mutations introduced into RB69 gp43 to induce PFA sensitivity (12). The arrows linking amino acids 365 and 478 represent the clashing Trp residues in the chimeric DNA pol. The small arrows point out mutations known to arise in drug-resistant HCMV strains. The mutation K805Q marks a PFA-sensitizing mutation, unlike the others, which are associated with PFA resistance.