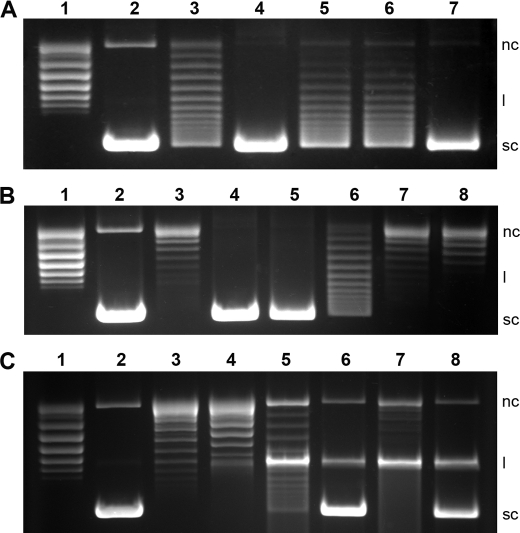
FIGURE 3.
Inhibition of DNA gyrase by ciprofloxacin/novobiocin and protection by QnrB1. A, QnrB1-mediated protection of gyrase (supercoiling activity) against ciprofloxacin. Lane 1, relaxed plasmid pBR322 alone; lane 2, relaxed pBR322 plus gyrase; lanes 3–7, relaxed pBR322, gyrase, and 2 μm ciprofloxacin in the presence of 20 nm QnrB1WT (lane 4), 2 μm loop-AB double deletion (lane 5), loop B deletion (lane 6), and loop A deletion (lane 7) mutants. B, concentration-dependent inactivation of gyrase (supercoiling activity) by ciprofloxacin in the presence of QnrB1. Lane 1, relaxed plasmid pBR322; lane 2, relaxed pBR322 plus gyrase; lane 3, relaxed pBR322, gyrase, and 10 μm ciprofloxacin in the absence of QnrB1WT; lanes 4–8, 2 (lane 4), 5 (lane 5), 10 (lane 6), 25 (lane 7), and 50 μm (lane 8) ciprofloxacin in the presence of 40 nm QnrB1WT. C, effect of QnrB1 on ATPase inhibition and cleavage complex stabilization. Lane 1, relaxed plasmid pBR322; lane 2, relaxed pBR322 plus gyrase; lanes 3 and 4, relaxed pBR322, gyrase and 2 μm novobiocin in the absence of QnrB1WT (lane 3) and in the presence of 200 nm QnrB1WT (lane 4); lanes 5–8, cleavage complex stabilization assays using 4 μm (lanes 5 and 6) and 16 μm ciprofloxacin (lanes 7 and 8) in the absence of QnrB1 (lanes 5 and 7), and in the presence of 200 nm QnrB1 (lanes 6 and 8), respectively. nc, l, and sc indicate the nicked circular, linear, and supercoiled forms of pBR322, respectively.