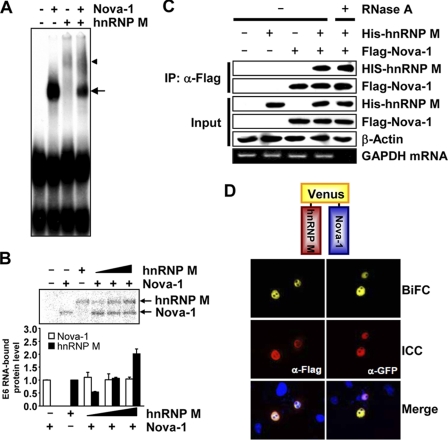
FIGURE 7.
Nova-1 interacts with hnRNP M. A, EMSA using Nova-1 (300 nm) and/or hnRNP M (300 nm) was carried out using radiolabeled E6. The arrow and arrowhead indicate specific binding of Nova-1 and hnRNP M to E6, respectively. B, UV cross-linking assays using Nova-1 (150 nm) and various amounts of hnRNP M (150 nm in the third lane and 75, 150, and 300 nm in the fourth, fifth, and sixth lanes, respectively) were performed on the D2R exon 6 probe. Cross-linked proteins were analyzed by SDS-PAGE, followed by autoradiography (upper panel). The band intensity of Nova-1 or hnRNP M was quantified using ImageQuant software (lower panel). Error bars represent the mean ± S.E. (n = 3). C, NIH3T3 cells were transiently transfected with FLAG-tagged Nova-1 and/or His-tagged hnRNP M. Total cell lysate was immunoprecipitated (IP) using anti-FLAG M2 affinity gel in the presence (+) or absence (−) of RNase A. Western blotting was performed with anti-hnRNP M, anti-FLAG, or anti-β-actin antibody to detect His-hnRNP M, FLAG-Nova-1, or β-actin, respectively (upper panels). GAPDH mRNA was analyzed using RT-PCR to demonstrate that RNase A digestion was complete (lower panel). D, HeLa cells were transiently transfected with VN-hnRNP M and VC-Nova-1. Twenty-four hours after transfection, the cells were fixed with paraformaldehyde; stained with anti-FLAG or anti-GFP antibody to visualize hnRNP M or Nova-1, respectively; and observed by confocal microscopy. Representative confocal images of cells are shown. BiFC, bimolecular fluorescence complementation; ICC, immunocytochemistry.