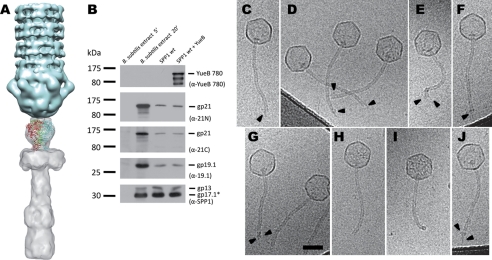
FIGURE 4.
Structural model of the SSP1 tail Tal extension to the SPP1 tip. A, docking of the gp21 C-terminal domain predicted structure into the SPP1 map of the tip. B, detection of gp19.1 and gp21 in extracts of B. subtilis infected with SPP1 (5 min and 20 min after infection) and in phage particles before and after DNA ejection triggered with YueB780. The amounts of sample applied to the gel lanes of both cellular extracts or of SPP1 particles were identical. DNA ejection from SPP1 phages was triggered by incubation with purified ectodomain receptor YueB780. Proteins were resolved in 10% SDS-PAGE gels and identified by Western blot. Two identical blots were probed sequentially with anti-21N, anti-19.1, and anti-SPP1 or with anti-21C and anti-YueB780 polyclonal antibodies, respectively. Note that YueB is not detected in B. subtilis extracts due to its low level of expression (40). The presence of the major capsid protein (gp13) and the major tail protein (gp17.1*) detected with anti-SPP1 serum is shown at the bottom. The positions of prestained molecular mass markers are indicated on the left (kilodaltons). Polyclonal antibodies used in each case are indicated within parentheses on the right. C–J, cryo-EM images of SPP1 tail ends after DNA ejection. SPP1 phages were incubated with the ectodomain receptor YueB780, inducing tip release and DNA ejection. Most phage capsids are entirely DNA-empty. Arrowheads (C–G) indicate structural heterogeneity of SPP1 tail ends. Sometimes (H and I), it is possible to observe well defined SPP1 tail ends, whereas rarely the YueB780 fiber can be see bound to the tail (J). Scale bar, 50 nm.