Abstract
Heparan sulfate (HS) and its highly modified form, 3-O-sulfated heparan sulfate (3-OS HS), contribute strongly to herpes simplex virus type-1 (HSV-1) infection in vitro. Here we report results from a random M13-phage display library screening to isolate 12-mer peptides that bind specifically to HS, 3-OS HS, and block HSV-1 entry. The screening identified representative candidates from two-different groups of anti-HS peptides with high positive charge densities. Group 1, represented by G1 peptide (LRSRTKIIRIRH), belongs to a class with alternating charges (XRXRXKXXRXRX), and group 2, represented by G2 peptide (MPRRRRIRRRQK), shows repetitive charges (XXRRRRXRRRXK). Viral entry and glycoprotein D binding assays together with fluorescent microscopy data indicated that both G1 and G2 were potent in blocking HSV-1 entry into primary cultures of human corneal fibroblasts and CHO-K1 cells transiently expressing different glycoprotein D receptors. Interestingly, G2 peptide isolated against 3-OS HS displayed wider ability to inhibit entry of clinically relevant strains of HSV-1 and some divergent members of herpesvirus family including cytomegalovirus and human herpesvirus-8. To identify functional residues within G1 and G2, we performed point mutations and alanine-scanning mutagenesis. Several arginine and a lysine residues were needed for anti-HSV-1 activity, suggesting the importance of the positively charged residues in virus-cell binding and virus-induced membrane fusion. In vivo administration of G1 or G2 peptide as a prophylactic eye drop completely blocked HSV-1 spread in the mouse cornea as evident by immunohistochemistry. This result also highlights an in vivo significance of HS and 3-OS HS during ocular herpes infection.
Keywords: Extracellular Matrix, Heparan Sulfate, Heparin, Heparin-binding Protein, Herpesvirus
Introduction
Heparan sulfate (HS)2 moieties present on cell surfaces provide attachment sites for many human and non-human pathogenic viruses including herpes simplex virus type-1 and -2 (HSV-1 and HSV-2) (1–4). Both wild-type and laboratory strains of HSV (5) bind HS. In addition to the attachment step, HSV-1 penetration into cells can also be mediated by 3-O-sulfated heparan sulfate (3-OS HS), which is produced after a rare enzymatic modification in HS catalyzed by 3-O-sulfortransferases (3-OSTs) (6–11). HSV envelope glycoproteins B and C bind HS and mediate virus attachment to cells (2, 3, 12). A third glycoprotein, gD, specifically recognizes 3-OS HS, and this interaction can facilitate fusion pore formation during viral entry (6, 7, 13). Despite the known importance of HS and 3-OS HS during HSV-1 infection in vitro (14, 15), not much is known about their significance during an actual infection in vivo.
The versatile ability of HS to bind a slue of microbes and participate in a variety of regulatory phenomena comes from its negatively charged nature and highly complex structure generated by enzymatic modifications (4, 16). Virtually all cells express HS as long un-branched chains often associated with protein cores commonly exemplified by syndecan and glypican families of HS proteoglycans (16). The parent HS chain, which contains repeating glucosamine and hexuronic acid dimers, can be 100–150 residues long and may contain multiple structural modifications. Most common among them is the addition of sulfate groups at various positions within the chain, which leads to the generation of specific motifs, making HS highly attractive for microbial adherence (4, 14, 15). Emerging evidence suggests that the role of HS in viral infection goes well beyond a role as a low specificity pre-attachment site. For instance, HS mediates HSV-1 transport on filopodia during surfing (17) and negatively regulates virus-induced membrane fusion (14). Likewise, for human papilloma virus, HS proteoglycans play a key role in the activation of immune response, an important aspect for both vaccine development and human papilloma virus pathogenesis (18). Similarly, HS expressed on spermatozoa plays a key role in the capture of human immunodeficiency virus (HIV) and its transmission to dendritic, macrophage, and T cells (19). 3-OS HS also plays a role in hepatitis B virus replication (20) and HS-binding peptides, or compounds can be successfully used for preventing genital human papilloma virus, HIV, and cytomegalovirus infection (21–24).
Despite the promise HS may show in anti-HSV therapy, virtually no products exist that target HS against human infections. Random peptide library screening has been used in the past to identify small peptides that specifically bind to unique immobilized protein targets and receptors expressed on cultured cells (25). The present study identifies and characterizes an anti-HS peptide, G1, and an anti-3-OS HS peptide, G2, that significantly inhibit HSV-1 infection and receptor-mediated cell-to-cell fusion in vitro. G2 peptide shows the extra promise of blocking infection of a divergent group of herpesviruses. The peptides also inhibit HSV-1 spread in a mouse model of corneal keratitis. To our knowledge, G1 and G2 peptides represent two novel classes of relatively small cationic peptides specifically isolated against HS and 3-OS HS, respectively, with strong herpesvirus entry-inhibiting activities. Inhibition of HSV-1 infection in the mouse cornea is the first demonstration of an in vivo significance of HS/3-OS HS in HSV-1 pathogenesis.
EXPERIMENTAL PROCEDURES
Selection of Phages against HS and 3-OS HS by Library Panning
A phage display library (PhDTM-12) expressing 12-mer peptides fused to a minor coat protein (pIII) of a non-lytic bacteriophage (M13) was purchased from New England Biolabs (Cambridge, MA). A purified form of HS isolated from bovine kidney was purchased from Sigma. Soluble 3-OS HS modified by 3-OST-3 was prepared as previously described (26). Screening of the phage display library was accomplished by an affinity selection (or bio-panning) process during which phage populations were selected for their ability to bind HS and 3-OS HS (modified by 3-OST-3). Both targets at a concentration of 10 μg/ml were used for overnight coating of wells of 96-well plates (Nalge Nunc International, Naperville, IL) in a humidifier chamber at 4 °C. The following day, the plates were blocked for 1 h at room temperature with 5 mg/ml bovine serum albumin in 0.1 m NaHCO3, pH 8.6, buffer. The plates were then washed 6 times with TBST (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% (v/v) Tween 20). The phage library was added to the plate at a concentration of 2 × 1011 in 100 μl in TBST. The plate was gently rocked for 1 h at room temperature. Unbound phages were removed by washing plates 10 times with 1 ml of TBST. Bound phages were eluted by adding 100 μl of Tris-HCl at pH 3.0. The eluate containing bound phages was removed, and the phages were amplified in Escherichia coli ER2738 bacteria and partially purified by polyethylene glycol precipitation. The binding, elution, and amplification steps were repeated using HS and 3-OST-3 modified HS as targets. Three rounds of selection were carried out to select for the specific binders. Low concentrations of detergent (Tween 20) in the early rounds resulted in high eluate titers, and the stringency was gradually increased with each round by raising Tween 20 concentration stepwise to a maximum of 0.5%. This allowed selection of high affinity binding phages. For final selection, the eluted phages were plaque-purified and tittered on soft agar plates.
Nucleotide Sequencing and Analysis
Automated nucleotide sequencing was performed to determine the sequences of the peptides encoded by the phages (Research Resource Center, University of Illinois at Chicago). The phage DNA was purified according to the manufacture's protocol using Qiagen mini-prep kit (Valencia, CA). DNA sequencing was initiated using ABI prism BigDye Terminator kit (Applied Biosystems, Foster City, CA) and the −96 gIII sequencing primer (New England Biolabs). Sequencing was performed on a Hitachi 3100 gene analyzer (Applied Biosystems), and the 36-nucleotide-long DNA region encoding the 12-mer peptides was identified and used for peptide synthesis. The synthetic peptides were resuspended at a concentration of 10 mm in phosphate buffer saline (PBS) at pH 7.4 and stored at −80 °C until use. The University of Illinois Research Resource Center was used for generating all the peptides and a few mutant peptides. The peptides for Ala-scanning mutagenesis were synthesized by GenScript Inc. The purity of the peptides was >95% as verified by high performance liquid chromatography, and the correct mass was confirmed by mass spectrometry.
Cell Culture and Viruses
Various cell types used in this study included wild-type Chinese hamster ovarian (CHO-K1), mutant CHO-745, and CHO-Iβ8 cells. In addition, primary cultures of human corneal fibroblasts (CFs), retinal pigment epithelial (RPE), human conjunctival epithelial (HCjE), Vero and HeLa cells were also used. CHO-K1cells were provided by P. G. Spear (Northwestern University, Chicago). HeLa cells were provided by B.P. Prabhakar (University of Illinois at Chicago). All CHO cell lines were grown in Ham's F-12 medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) and penicillin and streptomycin (Invitrogen). CF, HeLa, and RPE cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS and penicillin and streptomycin (27, 28), and Vero cells were grown in DMEM with 5% FBS and penicillin and streptomycin. Cultured HCjE cells provided by Dr. Illne Gipson (Harvard Medical School) were grown as described previously (29). The β-galactosidase expressing recombinant HSV-1(KOS)gL86 virus strain was provided by P. G. Spear. Similarly, HSV-1 strains used in this study (17, HfM, F, MP, and KOS) and HSV-2 G were also kindly provided by Dr. Spear. Green fluorescent protein (GFP) expressing HSV-1(K26GFP) and GFP expressing human herpesvirus-8 (HHV-8) were generous gifts, respectively, provided by Drs. Prashant Desai (Johns Hopkins University) (30) and J. Viera (University of Washington). The β-galactosidase-expressing recombinant cytomegalovirus (CMV) was obtained from ATCC.
HSV-1 Entry Assay
A previously described HSV-1 entry assay was used (6). CHO-K1 cells were grown in 6-well plates to subconfluence and then transfected with 2.5 μg of expression plasmids for gD receptors nectin-1 (pBG38), Herpesvirus Entry Mediator (pBec10), 3-OST-3 isoform (pDS43), or pCDNA3.1 (empty vector) using Lipofectamine (Invitrogen). At 16 h post-transfection, the cells were replated into 96-well dishes for preincubation with peptides at different concentrations for 60 min at room temperature. In parallel, natural target cells (HeLa, CF, and RPE) were also pretreated with the peptides for the same duration. In all cases the unbound peptides were removed after washing 3× with PBS. Thereafter, the cells were infected for 6 h with a recombinant virus, HSV-1(KOS)gL86, at multiple plaque-forming units (pfu), and β-galactosidase assays were performed using either a soluble substrate o-nitrophenyl-β-d-galactopyranoside (ONPG at 3.0 mg/ml; ImmunoPure, Pierce) or X-gal (Sigma). For the soluble substrate, the enzymatic activity was measured at 410 nm using a microplate reader (Spectra Max 190, Molecular Devices, Sunnyvale, CA). For the X-gal assay, cells were fixed (2% formaldehyde and 0.2% glutaraldehyde) and permeabilized (2 mm MgCl2, 0.01% deoxycholate, and 0.02% nonidet Nonidet P-40 (Sigma)). Finally, 1 ml of β-galactosidase reagent (1.0 mg/ml X-gal in ferricyanide buffer) was added to each well and incubated at 37 °C for 90 min before the cells were examined using bright field microscopy under the 20× objectives of the inverted microscope (Zeiss Axiovert 100M).
Fluorescent Microscopy of Viral Entry
Cultured monolayers of HeLa and CF cells (∼106 cells/well) were grown overnight in DMEM media containing 10% FBS on chamber slides (Lab-Tek). One pool of each cell type was pretreated with G1, G2, or a control peptide for 60 min. The cells were then infected with HSV-1(K26GFP) (30) at 50 pfu in serum-free media Opti-MEM (Invitrogen), and this was followed by fixation of cells at 90 min post-infection using fixative buffer (2% formaldehyde and 0.2% glutaraldehyde). The cells were then washed and permeabilized with 2 mm MgCl2, 0.01% deoxycholate and 0.02% Nonidet P-40 for 20 min. After rinsing, 10 nm rhodamine-conjugated phalloidin (Invitrogen) was added for F-actin staining at room temperature for 45 min. Finally, the cells were washed, and the images of labeled cells were acquired using a confocal microscope (Leica, Solms, Germany) and analyzed with MetaMorph software.
CMV Entry Assay
Natural target RPE cells were incubated with G1 and G2 peptides for 60 min at room temperature before the cells were infected with β-galactosidase expressing CMV (ATCC) for 8 h. β-Galactosidase assays were performed using either a soluble substrate o-nitrophenyl-β-d-galactopyranoside (ONPG at 3.0 mg/ml; ImmunoPure, Pierce) or X-gal (Sigma).
HHV-8 Infection Assay
HCjE cells grown in chamber slides (Labteck) or in a 96-well plate were pretreated with G1, G2, or control peptides for 60 min at room temperature followed by inoculation with recombinant rHHV-8.152, expressing the GFP (31). 48 h post-infection GFP-positive cells were visualized under microscope (Zeiss Axiovert 100M). HHV-8 infection was determined as relative fluorescence units using a GENios Pro plate reader (TECAN) at 480-nm excitation and 520-nm emission frequencies. Five measurements of negative control, positive control, and the test samples were performed. Data are expressed as the mean ± S.D.
HSV-1 Glycoprotein Induced Cell-to-cell Fusion Assay
In this experiment the CHO-K1 cells (grown in F-12 Ham's, Invitrogen), designated “effector” cells, were co-transfected with plasmids expressing four HSV-1(KOS) glycoproteins, pPEP98 (glycoprotein B), pPEP99 (glycoprotein D), pPEP100 (glycoprotein H), and pPEP101 (glycoprotein L) along with the plasmid pT7EMCLuc that expresses firefly luciferase gene under the T7 promoter (32). Wild-type CHO-K1 cells express cell surface HS but lack functional gD receptors and, therefore, are transiently transfected with HSV-1 entry receptors. Wild-type CHO-K1 cultured cells expressing HSV-1 entry receptors or naturally susceptible cells (human CFs) considered as “target cells” were co-transfected with pCAGT7 that expresses T7 RNA polymerase using chicken actin promoter and CMV enhancer (33). The untreated effector cells expressing pT7EMCLuc and HSV-1 essential glycoproteins and the target cells expressing gD receptors transfected with T7 RNA polymerase were used as the positive control. G2 or control peptide-treated target cells were then co-cultivated (1:1 ratio) for 18 h with effector cells for fusion. Activation of reporter luciferase gene as a measure of cell fusion was examined using reporter lysis assay (Promega) at 24 h post-mixing.
Flow Cytometry Analysis
Flow cytometry was performed to detect the effect of G2 peptide on GFP-HSV-1 binding to human CFs. Monolayers of ∼5 × 106 CFs were pretreated with G2 peptide for 60 min followed by incubation with GFP-expressing HSV-1(K26GFP) at 4 °C. Control peptide-treated or -untreated cells were similarly incubated with HSV-1 GFP virions. Uninfected CFs were used as the background negative control. GFP expression from the viral capsid on cell surface was examined by a flowcytometer (MoFlo).
Immunohistochemistry
BALB/c mice with pre-scarred corneal upper surface were treated with PBS-based eye drops containing 0.5 mm G1, G2, or control peptide followed by the inoculation of HSV-1(KOS). The mice were sacrificed after 4 and 7 days for HSV-1 detection. Immunohistochemistry was performed as described previously (29). Briefly, the tissue sections were hydrated with distilled water, and antigen retrieval was performed using DAKO Target Retrieval Solution 10× concentrate (DAKO, Carpinteria, CA). Nonspecific staining was blocked using an H2O2 solution for 10 min followed by a protein block for 10 min. Sections were incubated with HSV-1 gD-specific antiserum (1:100 dilution) at room temperature for 1 h followed by a 40-min incubation with the secondary antibody (HRP-conjugated goat anti-rabbit IgG, 1:500; Sigma). Expression of gD was detected using the DAKO Envision+ kit. Confocal and differential interference contrast image acquisition was conducted with an SB2-AOBS confocal microscope (Leica).
Statistics
The data presented are the means ± S.D. of triplicate measures of three or more experiments performed independently. Error bars represent the S.D. Statistical significance was calculated using Student's t test. A p value that was <0.05 was considered statistically significant.
RESULTS
Identification of HS and 3-OS HS-binding Peptides That Block HSV-1 Entry
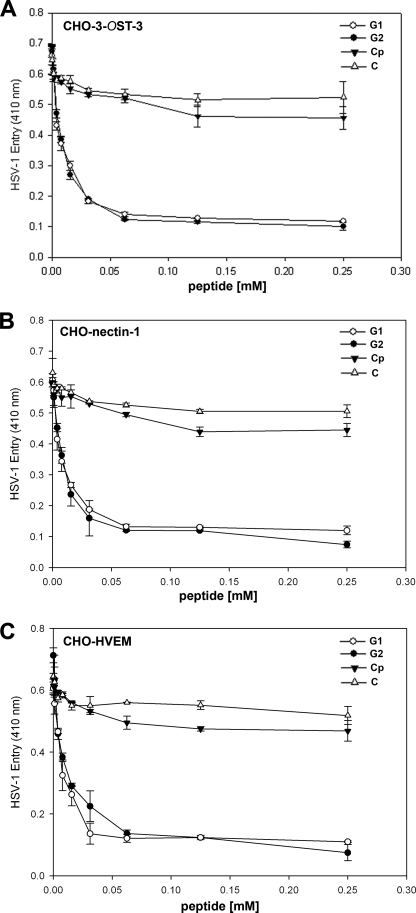
Three rounds of screening of phages from a 12-mer peptide phage display library resulted in the enrichment of phages that bound HS or 3-OS HS. Peptide sequences from individual plaques were deduced by determining the nucleotide sequences of the portion of the phage genome that encoded them. The predicted peptide sequences of about 200 plaques were determined and sorted into two groups on the basis of their targets. A frequently repeating peptide sequence from each group was subsequently selected for further characterization. The two most frequently isolated peptide sequences LRSRTKIIRIRH (called G1 for HS binding group 1) and MPRRRRIRRRQK (G2 for 3-OS HS binding group 2) were synthesized and examined for their abilities to inhibit HSV-1 entry. Both peptides were able to block HSV-1 entry into CHO-K1 cells expressing one of the three gD receptors, 3-OS HS, nectin-1, and HVEM. The blockage of entry occurred in a dose-dependent manner (Fig. 1) and it was independent of the gD receptor used. The concentration of each peptide that produces 50% of its maximum potential inhibitory effect (IC50) was determined. IC50 values ranged from 0.02 to 0.03 mm. A control phage bearing the sequence RVCGSIGKEVLG (designated control peptide (Cp)) did not inhibit HSV-1 entry, and none of the peptides showed any significant cytotoxicity (MTS assay (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium), Promega) at 5 mm or below concentrations. The highest concentration of the peptides in our experiments was kept at 0.5 mm.
FIGURE 1.
Inhibition of HSV-1 entry by 12-mer synthetic peptides is not specific to any particular gD receptor. G1 and G2 peptides were examined for their ability to block HSV-1 infection of 3-OST-3 (panel A)-, nectin-1 (panel B)-, and Herpesvirus Entry Mediator (panel C)-expressing CHO-K1 cells. Cells were preincubated with G1, G2, or control peptide (abbreviated as Cp) at the indicated concentrations in mm or mock-treated (abbreviated as C) with 1× phosphate saline buffer for 60 min at room temperature. After 60 min, a β-galactosidase-expressing recombinant virus HSV-1(KOS)gL86 (25 pfu/cell) virus was used for infection. After 6 h, the cells were washed, permeabilized, and incubated with ONPG substrate (3.0 mg/ml) for quantitation of β-galactosidase activity expressed from the input viral genome. The enzymatic activity was measured at an optical density of 410 nm. Each value shown is the mean of three or more determinations (±S.D.).
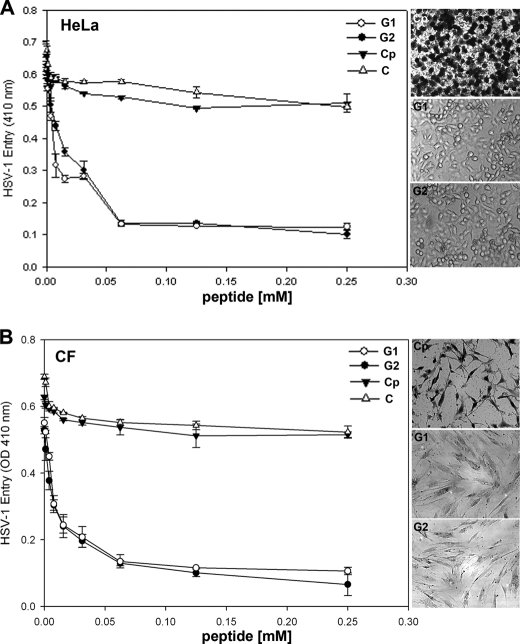
We next compared the abilities of the G1 and G2 peptides to block HSV-1 entry into natural target cells (HeLa and primary cultures of human CF). Again, a similar dosage response curve was generated when HeLa (Fig. 2A) or CF (Fig. 2B) cells were pretreated with G1 or G2 peptides during HSV-1 entry. The control peptide-treated cells had no effect on HSV-1 entry (Fig. 2). Use of insoluble blue cell assay (X-gal as the substrate for β-galactosidase) further confirmed that the peptides were effective in blocking infection of individual cells (Fig. 2, A and B, right panels). In virtually all cases, G2 peptide was slightly more effective in blocking entry than G1.
FIGURE 2.
G1 and G2 peptides block HSV-1 entry into human target cells. HeLa cells (panel A) and primary cultures of human CFs (panel B) were tested. Cells in 96-well plates were pretreated for 60 min with indicated mm concentrations of G1, G2, or Cp peptides. Mock-treated cells (abbreviated as C) served as a control. Pretreated cells were infected with a β galactosidase-expressing recombinant virus HSV-1(KOS)gL86 (25 pfu/cell) for 6 h. Viral entry was quantitated as described in Fig. 1. Confirmation of HSV-1 entry blocking activity of G1, G2, and Cp peptides on a per cell basis was obtained after cells were infected as described above followed by X-gal (1.0 mg/ml) staining (right panels), which yields an insoluble blue product upon hydrolysis by β-galactosidase expressed from the input viral genomes. Dark stained cells represent infected cell; uninfected cells stay transparent. Microscopy was performed using a 20 × objective of Zeiss Axiovert 100.
The Peptide Inhibitors Are Also Active against a Variety of HSV-1 Strains
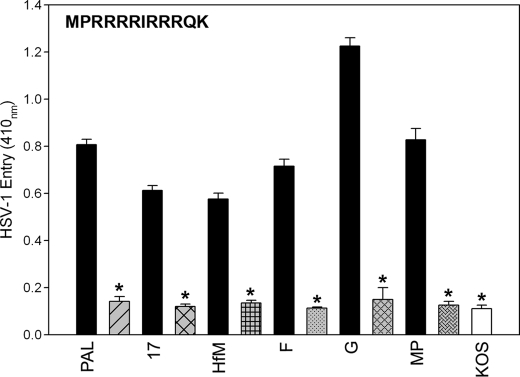
Next we evaluated the anti-HSV properties of G1 and G2 peptides against other commonly used strains (F and G, Hfem, MP, KOS, and 17) of HSV-1 and HSV-2 (34). Here we used 3-OST-3-expressing CHO Ig8 reporter cells that express β-galactosidase upon viral entry (35). The cells were preincubated with G1, G2, or Cp and subsequently infected with the viruses. The results from this experiment again showed that G2 (and similarly, G1; data not shown) blocked entry of various HSV-1 strains by 70–80% at 0.5 mm concentration (Fig. 3). Clearly, the inhibitory effect of G2 peptide was broad and not limited to any particular strains or serotypes.
FIGURE 3.
HSV-1 entry blocking activity of G2 peptide is not HSV-1 strain-specific. G2 (gray or white bars) or Cp control (black bars) at 0.5 mm concentration was incubated for 60 min at room temperature with reporter CHO-Ig8 cells that express β-galactosidase upon HSV-1 entry. After incubation, the cells were infected with HSV-1 (Pal, 17, Hfm, glycoprotein F, KOS, and MP) and HSV-2 (G) strains indicated above at 25 pfu/cell for 6 h at 37 °C. The viral entry blocking was measured by ONPG assay as described in Fig. 1.
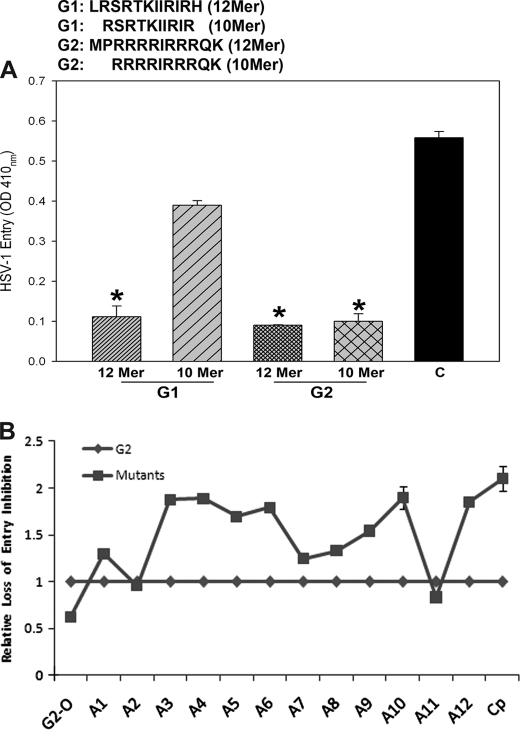
Structural Aspects of G1 and G2 Peptides: G2 Shows More Dependence on Charged Residues
To better understand the inhibitory potential of G1 and G2 peptides, we decided to make synthetic short versions (10-mer) lacking the terminal non-positively charged amino acids. In the case of G1, N-terminal Leu and C-terminal His residues were removed, and for G2, the N-terminal flanking residues (Met and Pro) were removed. We worked on a hypothesis that 12-mer peptides are too short to have any secondary or tertiary structure; therefore, their primary structure and most likely the presence of positively charged residues alone play a critical role during HSV-1 entry inhibition. As shown in Fig. 4A, the 10-mer version of G2 was very similar to the 12-mer in its ability to block HSV-1 entry into CFs. In contrast, the 10-mer version of G1 peptide almost completely lost its ability to block HSV-1 entry into the target cells. Thus, it is likely that terminal Leu and His are required for the anti-HSV-1 activity of G1. G2, on the other hand, relies more on its charged residues for its functional activity. To prove this point, we decided to perform alanine-scanning mutagenesis. It is commonly used to identify specific amino acid residues responsible for peptide function, stability, and conformation (36). Synthetic peptides were made whereby each residue of G2 was sequentially replaced with an Ala residue, and corresponding changes in G2 peptide were evaluated for the ability to affect viral entry (Fig. 4B). Quite interestingly, we found that the first four Arg residues were essential, whereas the middle two could be substituted with moderate loss of activity. The last Arg and Lys were also essential. Interestingly, the uncharged residues tolerated the substitutions well. Under similar experimental conditions, G2 oligomers were also examined, and it was evident that they blocked infection about 2-fold better than G1 (Fig. 4B). To conclude, our mutagenesis results suggest that the presence of positively charged residues plays an important role in HSV-1 entry blocking activity shown by G2.
FIGURE 4.
Deletion analysis and alanine-scanning mutagenesis reveal significance of positively charged residues in HSV-1 entry inhibition. A, synthetic 10-mer versions of G1 and G2 (shown above) were tested for blocking HSV-1(KOS)gL86 entry into cultured CF. After 6 h, the viral entry was measured by ONPG assay as described in Fig. 1. B, alanine-scanning mutagenesis was performed on G2. Twelve different peptides were synthesized replacing one residue at a time with an Ala, and their entry blocking activity was determined as described above. The location of Ala in the peptide is denoted by a number next to it, Cp represents the control peptide, and the oligomeric G2 is listed as G2 oligomers (G2-O). The figure shows relative loss of inhibitory potential upon substitution of a residue within G2 by an Ala. Activity of each peptide was normalized against the wild-type G2, which was kept at 1.00. Numbers higher than 1 show loss of activity, whereas a lower number represents gain of activity.
G2 Represents a Class of Broad Spectrum Anti-HS Peptides with Activity against Multiple Herpesviruses
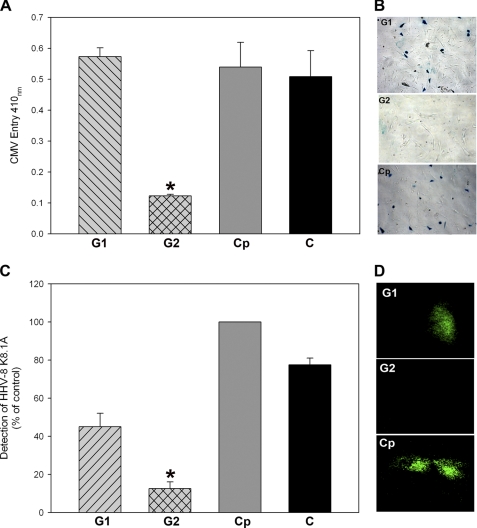
Usage of cell surface HS moieties during viral binding and entry is a unique feature that is shared by many infectious viruses including many herpesviruses (7). It is well documented that similar to HSV-1 (an α-herpesvirus), β- herpesvirus (CMV) and HHV-8 also use HS during cell entry and fusion (4, 7). Therefore, we made an optimistic attempt to test if G1 and G2 can also block infection of other members of herpesvirus family. To detect the peptide effect on viral entry, we used LacZ-expressing reporter CMV and GFP-expressing HHV-8 viruses (31) and employed their natural target cells for entry measurements. Surprisingly, G2 but not G1 peptide showed clear effects against CMV and HHV-8 (Fig. 5). As shown in Fig. 5A, G2 peptide was effective in blocking CMV entry into RPE cells, whereas G1 peptide had no effect. In essence, the effect of G1 was similar to the Cp or peptide-untreated cells, and the same pattern was repeated when the effects of the peptides were examined on a per cell basis by an X-gal assay (Fig. 5B). In parallel, the ability to block HHV-8 infection was examined in HCjE cells, a natural target for HHV-8 infection. Emission of fluorescence indicates virus infection. As shown in Fig. 5C, compared with G1- or Cp-treated cells G2-treated HcjE cells had relatively very low GFP expression. This suggests that G2 was able to block infection, and the result is statistically significant. Although G1 also demonstrated some lowering of fluorescence in the representative case shown in Fig. 5C, we did not find it statistically significant upon repeated experiments. This fact was also confirmed when individual cells were examined under a fluorescence microscope. Cells treated with G2 did not show fluorescence originating from GFP virus replication (Fig. 5D). However, the fluorescence was more easily seen with G1- or Cp-treated cells. The results suggest that G2 is more effective that G1 in blocking entry of divergent herpesviruses. G1 may have some activity, but the virus can possibly overcome it easily.
FIGURE 5.
G2 also blocks entry of representative members of β- and γ-herpesvirus subfamilies (CMV and HHV-8). A, a monolayer of cultured RPE cells grown in a 96-well plate was pretreated with G1, G2, or Cp at 0.5 mm concentration. A mock-treated population of RPE cells served as the positive control (abbreviated as C). After 60 min of incubation at room temperature, the cells were infected with β-galactosidase expressing CMV reporter virus. After 8 h, viral entry was measured as described for Fig. 1. B, effect of CMV entry blocking activity of G1, G2, or Cp peptide for individual RPE cells was determined by X-gal staining, which yields an insoluble blue product upon hydrolysis by β-galactosidase. Individual cells were examined using Zeiss Axiovert 100 microscope at 20× magnification. Infected cells turn blue. C, HCjE cells preincubated with G1, G2, or Cp were infected with HHV-8 virions for 48 h at 37 °C. After incubation, the cells were washed thoroughly to remove unbound viruses. GFP-expression of HHV-8 was quantitated by determining relative fluorescence units using a 96-well fluorescence reader (TECAN). The data shown are the means of triplicate measures and are representative of three independent experiments. D, viral replication in HCjE cells were visualized under fluorescent microscope (Zeiss Axiovert 100) in cells that were pretreated with G1, G2, or Cp as described above. Asterisks indicate significant difference from controls and/or treatments (p < 0.05, t test), and error bars represent S.D.
Mechanism for HSV-1 Entry Inhibition by the Peptides
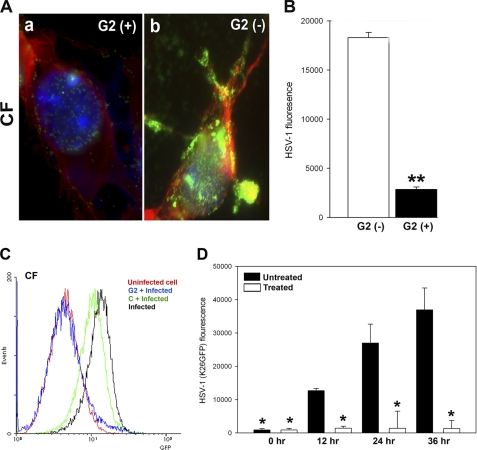
Because G2 peptide showed significant blocking activity against multiple herpesviruses, we focused our attention on G2 to determine how it may function to block infection. One possible explanation is based on its ability to block the sites on HS that may otherwise bind the virus. To examine this, we made use of a GFP-tagged HSV-1(K26GFP) virus. Cultured CF were preincubated with 0.5 mm G2 peptide or Cp and then infected with the GFP virus. Cells were fixed at 60 min post-infection and stained for F-actin (red) and the nucleus (DAPI, blue). It was evident that G2-treated cells resisted virus attachment (Fig. 6A, green) compared with the control peptide-treated cells. To examine this effect on a population of 105 cells, GFP intensity as an indicator of virus binding was measured after incubation with the virus in cold and rigorous washing of the cells afterward (Fig. 6B). Clearly, the binding was significantly higher in Cp-treated compared with G2 peptide-treated cells. These results together with those of the entry assay and visualization of GFP-tagged HSV-1 suggest that pretreatment of cells with G2 peptide significantly blocks virus attachment and, hence, the entry of HSV-1 into natural target cells.
FIGURE 6.
G2 functions by preventing HSV-1 attachment to cells, which results in loss of binding and viral replication. A, GFP-expressing HSV-1(K26GFP) binding to CF in the presence and absence of G2 peptides was examined by fluorescence microscopy. CF were grown in collagen-coated chamber slides and incubated at room temperature for 60 min with G2 (+) or control Cp (G2 (−)) peptide. This was followed by the incubation of the cells in cold with GFP-expressing HSV-1(K26GFP) for 30 min and washing of unbound virioins with PBS. Cells were fixed, stained with phalloidin for F-actin (red) and DAPI for nuclei (blue), and examined by a fluorescence microscope (Leica, SP2). The presence of the virus is shown in green. B, relative virus binding to CF was estimated by fluorescence measurements. Cultured CF were preincubated with G2 and Cp for 60 min before ice-cold incubation with GFP-expressing HSV-1 virus for 30 min. Cells were washed 3 times, and viruses remaining on cell surfaces were assayed for GFP fluorescent intensity using a fluorescence reader (Tecan). C, GFP-expressing HSV-1(K26GFP) intensity as a surrogate for virus binding was quantified in presence G2 or control peptide (abbreviated as C) by flow cytometry. The cell/virus incubation was performed as described above. D, G2 peptides block HSV-1 replication into cultured human CFs. Cultured CF were preincubated with G2 or mock-treated (Cp) before infection with HSV-1(K26GFP) virus for 6 h. Viral replications in CF were quantified 0–36 h post-infection by measuring GFP fluorescent intensity using a fluorescence reader (Tecan). The data shown are the means of triplicate measures and are representative of three independent experiments. Asterisks indicate significant difference from other treatments (p < 0.01, t test); error bars represent S.D.
G2 Peptide Acts by Inhibiting HSV-1 Binding to HS
Next, we evaluated the effect of G2 on virus binding by flow cytometry. We reasoned that a GFP reporter virus binding on cells pretreated with G2 will be significantly reduced, and this should be detectable by flow cytometry. Primary cultures of human CF pretreated with G2 peptide or control peptide (C) were analyzed for HSV-1(K26GFP) binding. The peptide-untreated CF incubated with the virus served as a positive control, whereas the uninfected peptide was kept as a negative background control. Our results indicate that the virus failed to bind G2 peptide-treated CF (Fig. 6C). For unknown reasons, the control peptide also showed some inhibition of binding, but the effect of G2 was far more evident. This result confirms that G2 has the ability to block virus attachment to cells. To further confirm that blocking of the attachment means no virus replication either, we monitored fluorescence over time as a measure of K26GFP (30) virus production in G2-treated and mock-treated cells. The GFP intensity (representing virus production) increased significantly over time (Fig. 6D) in mock-treated cells but not when the cells were treated with G2. Thus, our result suggests that G2 can block virus binding to a level that no replication can be detected.
Pretreatment of G2 Peptide to the Target Cell Significantly Affects Cell-to-cell Fusion and Viral Spread
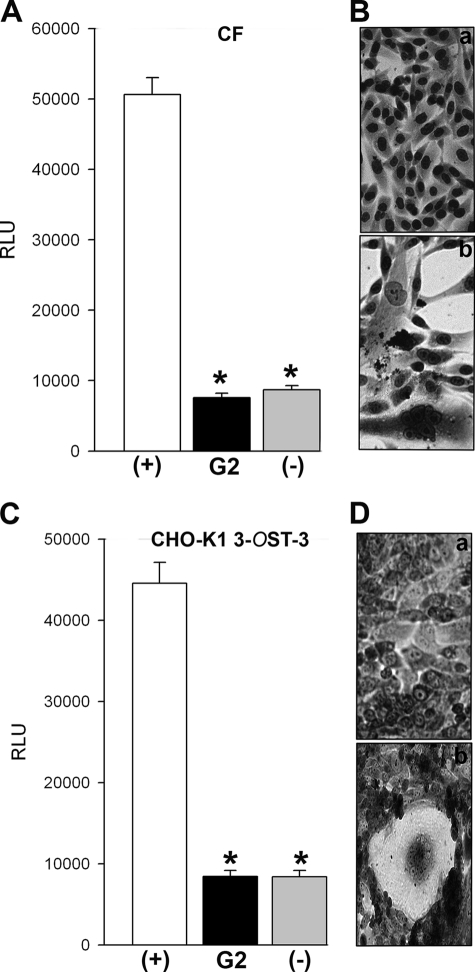
Because G2 was isolated against 3-OS HS, which can mediate viral penetration by promoting membrane fusion (13), we decided to test whether G2 also possesses the ability to block the membrane fusion induced by HSV-1 glycoproteins (32). The same membrane fusion is used during polykaryocytes formation and cell-to-cell spread (33, 37). 3-OST-3 expressing CHO-K1 cells and primary cultures of human CF were preincubated with G2 peptide followed by co-culture with effector CHO-K1 cells expressing HSV-1 glycoproteins. The membrane fusion that ensues upon co-culturing the cells can be estimated by a luciferase-based reporter assay (32). Likewise, the polykaryocytes formation can be visualized by Giemsa staining. Evidently, a prior treatment with G2 was very effective in blocking membrane fusion (Fig. 7, A and C), and this ability also translated into the loss of syncytia formation (Fig. 7, Ba and Da) compared with mock-treated cells (Fig. 7, Bb and Db). These findings indicate that G2 not only blocks attachment, but it can also inhibit viral penetration by blocking membrane fusion.
FIGURE 7.
Effect of G2 on HSV-1 glycoprotein induced cell-to-cell fusion and spread. A, the effector CHO-K1 cells expressing HSV-1 glycoproteins (B, D, H–L, and T7 polymerase) were preincubated with G2 peptide (black bar) or 1 × PBS (white bar) (+) for 90 min. Control effector cells (T7 polymerase and glycoproteins gD, H–L only) (−) were also preincubated with G2 for the same duration. The effector cells were then mixed with primary cultures of human corneal fibroblasts (panels A and B) or 3-OST-3-expressing CHO-K1 cells (panels C and D) transfected with luciferase gene under T7 control. Membrane fusion as a surrogate for viral spread was detected by monitoring luciferase activity (panels A and C). Relative luciferase units (RLUs) were determined using a Sirius luminometer (Berthold detection systems). Error bars represent S.D. *, p < 0.05, one way analysis of variance. Microscopic images of Giemsa (Fluka)-stained polykaryocytes show the preventative effect of G2s on cell fusion (panels B and D). Shown above are 40× magnified photographs of cells undergoing membrane fusion (Zeiss Axiovert 200).
G1 and G2 Peptides Show Protective Effects against HSV-1 Infection of the Mouse Cornea
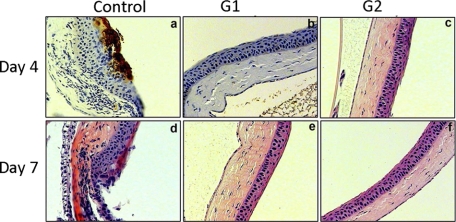
Finally, we tested the abilities of G1 and G2 peptides against HSV-1 infection in a mouse model. We targeted mouse cornea for two reasons. First, the cornea is known to express many gD receptors including 3-OS HS (27, 33). Second, cornea is an attractive target for HSV-1 infection leading to the development of herpetic stromal keratitis, a potential blinding disease common in developed countries including the United States (38). Immunohistochemistry was used to locate HSV-1 gD expression in the cornea pretreated with either a control peptide, G1, or G2 followed by HSV-1 infection. In sections of cornea of mice euthanized at the fourth and seventh days after pretreatment with Cp peptide (control) followed by virus inoculation demonstrated severe chronic inflammation combined with significant staining for HSV-1 gD on day 4 (Fig. 8, panel a). HSV-1 staining was gone by day 7, which is typical with normal mice; however, damage to the corneal epithelium was still evident (Fig. 8, panel b). In contrast, virtually no HSV-1 protein expression was detected in the corneas treated with G1 or G2 peptide, and the epithelium remained intact at both 4 and 7 days post-infection (Fig. 8, panels b, c, e, and f). These findings prove two important points that anti-HS and anti-3-OS HS peptides show strong promise as anti-HSV prophylactic agents and that HS is an important co-receptor for an ocular HSV-1 infection in vivo.
FIGURE 8.
G1 or G2 effectively block infection in a mouse model of corneal keratitis. 100 μl of G1, G2, or Cp (control) peptide at 0.5 mm concentration was poured into the mouse cornea as a prophylactic “eye drop” followed by an infection with HSV-1(KOS) at 106 pfu. At 4 or 7 days post-infection, immunohistochemistry was performed using anti-HSV-1 gD polyclonal antibody. Staining for HSV-1 is shown in brown.
DISCUSSION
In this report we used phage display peptide library screening as a way to identify small and easily synthesizable peptides that specifically bind HS and/or 3-OS HS. The two most frequently isolated peptides, one against commercially found HS (G1) and the other against specifically modified 3-OS HS (G2), were investigated for their ability to block HSV-1 entry and spread. The entry blocking activity of the peptides was not dependent on any particular gD receptor, virus strain, or cell type used. We also found that the peptides function by interfering with the binding of HSV-1 to cells. To our understanding ours is the first demonstration of small 12-mer peptides specifically screened against HS/3-OS HS that block virus attachment and thereby inhibit viral entry.
Our library screening resulted in isolation of 200 HS binding phages. The peptides encoded by the phages were enriched in basic amino acid residues, and they were classified into two major groups. In The first group, represented by G1, the sequences had basic amino acids after every residue (XRXRXKXXRXRX). In the second group, represented by G2, repeats of basic amino acids (XXRRRRXRRRXK) were observed. This is very interesting because G2 was isolated against 3-OS HS, and alanine-scanning mutagenesis suggested that the repeats, especially the first one, were essential for entry inhibition (Fig. 4B). Unlike G1, G2 also blocks the entry of two divergent herpesviruses, CMV and HHV-8 (7). In addition, we also found that G2 blocks membrane fusion as well, which means the peptide can interfere with gD interaction with its receptor, 3-OS HS (39–41). Among the structural differences between G1 and G2, it appears that G2 shows more dependence on the positively charged residues than G1, which is probably also dependent on the presence of a Lys residue at the N terminus. In general, arginine has been found important for charge-charge interaction with HS (42).
The ability of our peptides to act specifically on HS/3-OS HS is highly significant for many additional reasons. First, HS is widely expressed on all cells and tissues, and it is known to regulate many important biological phenomena (16, 43). Our peptides can be used as probes to study HS functions. Second, many times HS moieties are up-regulated especially during pathological conditions, and they themselves may also contribute to inflammation (16). Our peptides may find usage in blocking pathological effects of HS and regulating inflammation as well. Third, a complex enzymatic regulation of HS chain gives HS an equally complex ability (and affinity) to bind many proteins to perform new functions (16, 43). The 3-OS HS binding peptide may be used as a probe or a diagnostic tool to assess structural alterations within HS or its turnover on cell surfaces. Overall, the peptides cannot only be used against viral infections but also for many additional unrelated applications. Even for anti-viral activity, many unrelated viruses bind HS (4), and it is quite possible that G2 may have the ability to block infection of many additional viruses, a possibility that requires further scrutiny.
One additional and a very important use of the peptides is to demonstrate the significance of HS during an actual HSV-1 infection in vivo. Although HS has been well studied as an attachment receptor (7), very limited information is available on its usage in vivo. Our experiments using a mouse corneal infection model not only show the efficacy of the peptides in blocking infection in vivo, but it also shows that HS is an important HSV-1 coreceptor, not only for cultured cells but also for the cells in vivo (Fig. 8). Mice treated with either G1 or G2 did not show any obvious symptoms of infection, and likewise, a key viral protein was undetectable. Future experiments will determine whether a prior treatment with G1 and G2 can also stop the virus from establishing a latent infection or not. Likewise, it will be interesting to see the effects G1 and G2 may have on an existing HSV-1 infection (therapeutic potential) and effects of G2 on other viral infections. It will also be very important to determine the fate of the peptide(s) once it is bound to the cell surface HS. We have been unable to detect any redistribution of HS on cell surfaces; however, it is quite possible that the peptide is internalized, and it may eventually be delivered to the nucleus.3 In that case the peptide(s) may provide an interesting vehicle for gene and protein delivery to the nucleus. In any case, interfering with virus entry is an attractive prophylactic strategy that may have the ability to control virus infection and prevent transmission. We have provided the proof of principle for a possible clinical benefit of our peptides.
Acknowledgments
We thank Drs. Beatrice Yue (University of Illinois at Chicago) and Patricia Spear (Northwestern University, Chicago) for reagents and Drs. Christian Clement, Perry Scanlan, and Myung Jin Oh for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 AI057860 (to D. S.) and AI050050 (to J. L.) and Core Grant EY001792 (to the University of Illinois at Chicago). Part of this work was made possible by an award from the Glaucoma Foundation (to D. S.).
V. Tiwari, J. Liu, T. Valyi-Nagy, and D. Shukla, unpublished results.
- HS
- heparan sulfate
- 3-OS HS
- 3-O-sulfated heparan sulfate
- gD
- glycoprotein D
- CF
- corneal fibroblasts
- HHV-8
- human herpesvirus-8
- RPE
- retinal pigment epithelial
- HCjE
- human conjunctival epithelial
- pfu
- plaque forming unit
- ONPG
- o-nitrophenyl-β-d-galactopyranoside
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside
- Cp
- control peptide
- 3-OST
- 3-O-sulfortransferase
- 3-OST-3
- 3-O-sulfotransferase-3.
REFERENCES
- 1. Lycke E., Johansson M., Svennerholm B., Lindahl U. (1991) J. Gen. Virol. 72, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 2. Shieh M. T., WuDunn D., Montgomery R. I., Esko J. D., Spear P. G. (1992) J. Cell Biol. 116, 1273–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. WuDunn D., Spear P. G. (1989) J. Virol. 63, 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu J., Thorp S. C. (2002) Med. Res. Rev. 22, 1–25 [DOI] [PubMed] [Google Scholar]
- 5. Trybala E., Roth A., Johansson M., Liljeqvist J. A., Rekabdar E., Larm O., Bergström T. (2002) Virology 302, 413–419 [DOI] [PubMed] [Google Scholar]
- 6. Shukla D., Liu J., Blaiklock P., Shworak N. W., Bai X., Esko J. D., Cohen G. H., Eisenberg R. J., Rosenberg R. D., Spear P. G. (1999) Cell 99, 13–22 [DOI] [PubMed] [Google Scholar]
- 7. Shukla D., Spear P. G. (2001) J. Clin. Invest. 108, 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xia G., Chen J., Tiwari V., Ju W., Li. J. P., Malmstrom A., Shukla D., Liu J. (2002) J. Biol. Chem. 277, 37912–37919 [DOI] [PubMed] [Google Scholar]
- 9. Tiwari V., O'Donnell C. D., Oh M. J., Valyi-Nagy T., Shukla D. (2005) Biochem. Biophys. Res. Commun. 338, 930–937 [DOI] [PubMed] [Google Scholar]
- 10. Xu D., Tiwari V., Xia G., Clement C., Shukla D., Liu J. (2005) Biochem. J. 385, 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O'Donnell C. D., Tiwari V., Oh M. J., Shukla D. (2006) Virology 346, 452–459 [DOI] [PubMed] [Google Scholar]
- 12. Herold B. C., WuDunn D., Soltys N., Spear P. G. (1991) J. Virol. 65, 1090–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tiwari V., Clement C., Duncan M. B., Chen J., Liu J., Shukla D. (2004) J. Gen. Virol. 85, 805–809 [DOI] [PubMed] [Google Scholar]
- 14. O'Donnell C. D., Shukla D. (2009) J. Biol. Chem. 284, 29654–29665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O'Donnell C. D., Kovacs M., Akhtar J., Valyi-Nagy T., Shukla D. (2010) Virology 397, 389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Esko J. D., Lindahl U. (2001) J. Clin. Invest. 108, 169–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oh M. J., Akhtar J., Desai P., Shukla D. (2010) Biochem. Biophys. Res. Commun. 391, 176–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Witte L., Zoughlami Y., Aengeneyndt B., David G., van Kooyk Y., Gissmann L., Geijtenbeek T. B. (2007) Immunobiology 212, 679–691 [DOI] [PubMed] [Google Scholar]
- 19. Ceballos A., Remes, Lenicov F., Sabatté J., Rodríguez, Rodrígues C., Cabrini M., Jancic C., Raiden S., Donaldson M., Agustín, Pasqualini R., Jr., Marin-Briggiler C., Vazquez-Levin M., Capani F., Amigorena S., Geffner J. (2009) J. Exp. Med. 206, 2717–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Z., Liu X., Chen J., Su H., Luo Q., Ye J., Tang N., Zhang W., Chen W., Ko B. C., Huang A. (2010) Virology 406, 280–285 [DOI] [PubMed] [Google Scholar]
- 21. Baleux F., Loureiro-Morais L., Hersant Y., Clayette P., Arenzana-Seisdedos F., Bonnaffé D., Lortat-Jacob H. (2009) Nat. Chem. Biol. 5, 743–748 [DOI] [PubMed] [Google Scholar]
- 22. Donalisio M., Rusnati M., Civra A., Bugatti A., Allemand D., Pirri G., Giuliani A., Landolfo S., Lembo D. (2010) Antimicrob. Agents Chemother. 54, 4290–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luganini A., Giuliani A., Pirri G., Pizzuto L., Landolfo S., Gribaudo G. (2010) Antiviral Res. 85, 532–540 [DOI] [PubMed] [Google Scholar]
- 24. Copeland R., Balasubramaniam A., Tiwari V., Zhang F., Bridges A., Linhardt R. J., Shukla D., Liu J. (2008) Biochemistry 47, 5774–5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mullen L. M., Nair S. P., Ward J. M., Rycroft A. N., Henderson B. (2006) Trends Microbiol. 14, 141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tiwari V., O'donnell C., Copeland R. J., Scarlett T., Liu J., Shukla D. (2007) J. Gen. Virol. 88, 1075–1079 [DOI] [PubMed] [Google Scholar]
- 27. Tiwari V., Clement C., Xu D., Valyi-Nagy T., Yue B. Y., Liu J., Shukla D. (2006) J. Virol. 80, 8970–8980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tiwari V., Oh M. J., Kovacs M., Shukla S. Y., Valyi-Nagy T., Shukla D. (2008) FEBS J. 275, 5272–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Akhtar J., Tiwari V., Oh M. J., Kovacs M., Jani A., Kovacs S. K., Valyi-Nagy T., Shukla D. (2008) Invest. Ophthalmol. Vis. Sci. 49, 4026–4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Desai P., Person S. (1998) J. Virol. 72, 7563–7568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vieira J., O'Hearn P., Kimball L., Chandran B., Corey L. (2001) J. Virol. 75, 1378–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pertel P. E., Fridberg A., Parish M. L., Spear P. G. (2001) Virology 279, 313–324 [DOI] [PubMed] [Google Scholar]
- 33. Tiwari V., ten Dam G. B., Yue B. Y., van Kuppevelt T. H., Shukla D. (2007) FEBS Lett. 581, 4468–4472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dean H. J., Terhune S. S., Shieh M. T., Susmarski N., Spear P. G. (1994) Virology 199, 67–80 [DOI] [PubMed] [Google Scholar]
- 35. Montgomery R. I., Warner M. S., Lum B. J., Spear P. G. (1996) Cell 87, 427–436 [DOI] [PubMed] [Google Scholar]
- 36. O'Nuallain B., Allen A., Ataman D., Weiss D. T., Solomon A., Wall J. S. (2007) Biochemistry 46, 13049–13058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tiwari V., Darmani N. A., Thrush G. R., Shukla D. (2009) Biochem. Biophys. Res. Commun. 390, 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liesegang T. J. (2001) Cornea 20, 1–13 [DOI] [PubMed] [Google Scholar]
- 39. Akhtar J., Shukla D. (2009) FEBS J. 276, 7228–7236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Clement C., Tiwari V., Scanlan P. M., Valyi-Nagy T., Yue B. Y., Shukla D. (2006) J. Cell Biol. 174, 1009–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Spear P. G., Longnecker R. (2003) J. Virol. 77, 10179–10185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Knappe M., Bodevin S., Selinka H. C., Spillmann D., Streeck R. E., Chen X. S., Lindahl U., Sapp M. (2007) J. Biol. Chem. 282, 27913–27922 [DOI] [PubMed] [Google Scholar]
- 43. Bishop J. R., Schuksz M., Esko J. D. (2007) Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]