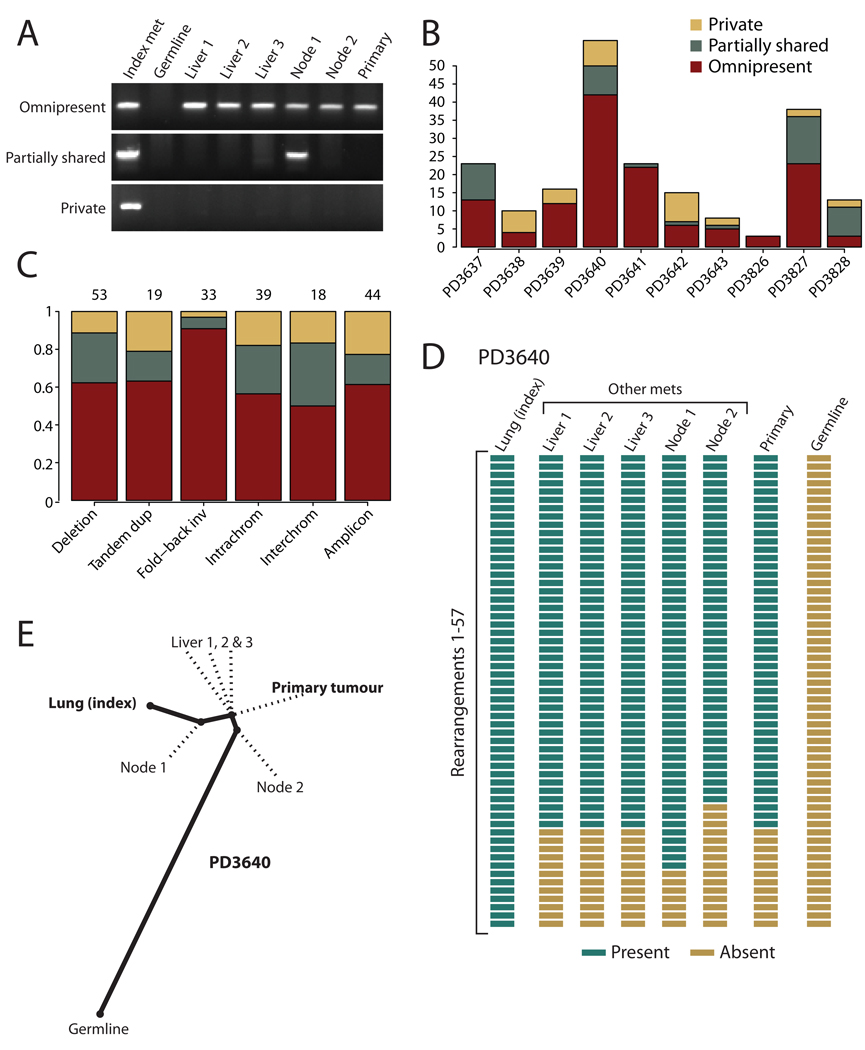
Figure 2.
Phylogenetic relationships of different metastases within a patient. (A) PCR genotyping of three rearrangements across DNA from the index metastasis sequenced, other metastases from the same patient, the primary tumour and germline tissue. Somatic rearrangements may be present in all cancer samples but not the germline (omnipresent); present in some but not all metastases (partially shared); or present just in the index metastasis sequenced (private). (B) Inter-individual differences in the proportions of rearrangements that are omnipresent across metastases, partially shared by some but not all lesions or are private to the index metastasis sequenced. (C) Patterns across six broad categories of rearrangement in the proportions of variants that are omnipresent across metastases, partially shared by some but not all lesions or are private to the index metastasis sequenced. The numbers of rearrangements in each category are shown at the top. The difference in proportions between fold-back inversions and the other categories was statistically significant (p=0.003). (D) Genotyping of 57 rearrangements in PD3640 shows a coherent, nested structure, with 42 found in all metastases and the primary tumour, 7 found uniquely in the index tumour and 8 partially shared by some but not all metastases. (E) The nested structure of rearrangements defines a phylogenetic tree of relationships among the metastases and primary tumour. The length of heavy black lines is proportional to the genetic distance between nodes. Dotted lines delineate the departure points of other, unsequenced lesions from the lineage between the germline genome and that of the index metastasis.