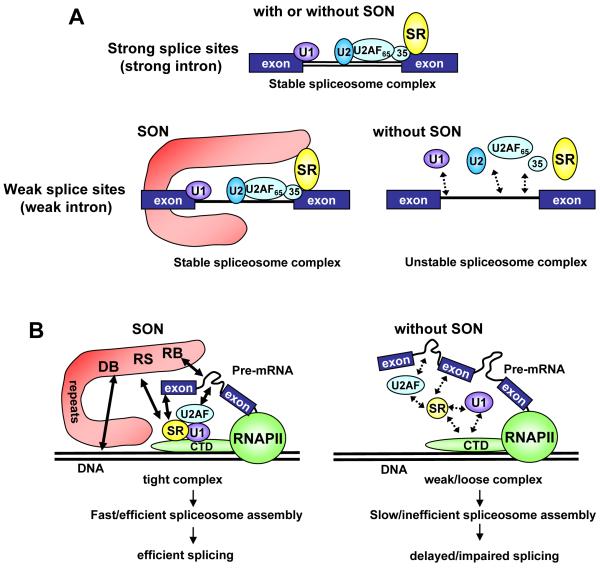
Figure 7. Proposed models for the role of SON as a splicing co-factor.
(A) If splice sites are strong (which can be due to optimal sequences in the intron), interactions between pre-mRNA and spliceosome components are strong and stable. Therefore, spliceosome complex is formed for splicing regardless presence or absence of SON. When splice sites are weak (due to suboptimal sequences), spliceosome components form weak and unstable interactions (represented by dotted lines) with pre-mRNA in the absence of SON, while stable interactions can be assured by presence of SON. The process may involve interaction(s) between SON and other critical splicing factors, including SR proteins.
(B) A model for the role of SON as a co-factor in efficient transcription-splicing coupling. During transcription in wild type cells (left), SON interacts with DNA, RNA, SR proteins and other early spliceosome components through its DNA-binding domain (DB), RNA-binding motifs (RB) and RS domain (RS), thereby facilitating recruitment of early spliceosome components to the CTD of RNAP II. The long and unique amino acid repeats in SON may help this protein stretch and make contact with multiple components. Such organization and connection by SON may help efficient and immediate spliceosome assembly on the nascent pre-mRNA with a weak splice site, resulting in efficient splicing. In the absence of SON (right), SR proteins and other early spliceosome components are not efficiently recruited to the CTD of RNAP II, and DNA, RNA and proteins are not closely associated with each other. Therefore, co-transcriptional spliceosome assembly is not efficient, and splice site recognition/selection on a weak splice site is not accurate, resulting in delayed or impaired splicing.