Abstract
Sequencing the coding regions, the exome, of the human genome is one of the major current strategies to identify low frequency and rare variants associated with human disease traits. So far, the most widely used commercial exome capture reagents have mainly targeted the consensus coding sequence (CCDS) database. We report the design of an extended set of targets for capturing the complete human exome, based on annotation from the GENCODE consortium. The extended set covers an additional 5594 genes and 10.3 Mb compared with the current CCDS-based sets. The additional regions include potential disease genes previously inaccessible to exome resequencing studies, such as 43 genes linked to ion channel activity and 70 genes linked to protein kinase activity. In total, the new GENCODE exome set developed here covers 47.9 Mb and performed well in sequence capture experiments. In the sample set used in this study, we identified over 5000 SNP variants more in the GENCODE exome target (24%) than in the CCDS-based exome sequencing.
Keywords: human exome, resequencing, GENCODE
Introduction
Exome resequencing is increasingly becoming a standard tool for the discovery of genes underlying rare monogenic disease and the discovery of coding variants associated with common disease.1, 2, 3 Although the cost of whole-genome sequencing has fallen dramatically over the last 2–3 years, it is still too expensive to be a useful approach for the identification of variants associated with different phenotypes in large cohorts. However, the combination of ‘second-generation' sequencing technologies (reviewed in refs 4,5) with robust and efficient methods of sequence capture6, 7, 8, 9 has enabled the widespread targeting of the exome.
Exome resequencing studies are, however, currently being performed using designs based on an incomplete exome, and consequently many medically relevant genes are not being screened in ongoing large-scale disease studies. The two most widely used commercial kits for capturing the exome target exons from genes in the consensus coding sequence (CCDS10) consortium database, in addition to a selection of miRNAs and non-coding RNAs, are NimbleGen Sequence Capture 2.1M Human Exome Array, http://www.nimblegen.com/products/seqcap/ and Agilent SureSelect Human All Exon Kit, http://www.genomics.agilent.com. Although the collaborative effort behind the CCDS database has provided a high-quality set of consistently annotated protein-coding regions, there are still many annotated genes, with solid evidence of transcription, that are not yet part of this set. In addition, only 21% of CCDS genes have an alternative spliced variant annotated. To address this shortcoming, we have designed and experimentally tested a more complete set of target regions for the human exome, based on the GENCODE annotation11 (release 2). The GENCODE collaboration is part of the Encode project and responsible for the annotation and experimental validation of gene loci on the human genome.
Materials and methods
Bioinformatic bait design
To generate the coordinates for the GENCODE exome, we extracted the coordinates for a total of 288 654 unique exons from 46 275 transcripts of 20 921 Ensembl12 protein-coding genes (release 53) and 33 621 transcripts of 13 772 manually annotated protein-coding genes (HAVANA,13 database version February 2009), together with an additional 1635 miRNA genes (Ensembl/miRBase). If the coordinates of any of these exons overlapped by one or more base pairs, regardless of strand, the overlapping exons were clustered together into expressed cluster regions (ECRs). A 10 bp flank was added on both sides of each ECR. Any ECR that now overlapped as a result of this flank by at least 1 bp was merged. This resulted in 207 108 ECRs, covering ∼39.3 Mb as the design target (35.2 Mb of exonic sequence plus 4.1 Mb of flanking sequence). The coordinates of these regions were used for bait design. The baits were created using the Agilent SureSelect design algorithm in three rounds of design using RepeatMasker- and WindowMasker-defined repeats in an attempt to avoid repetitive regions as well as to increase coverage of the target exons. Each successive round of design was more permissive of repeat overlap (0, 20 and 40 bp). After sequencing, the underperforming baits were boosted at specific ratios to even out the coverage across all targets. Depending on the placement of baits relative to repeat regions, the boosting was done either by direct replication or by shifting the booster bait either up or downstream by 30 bp. It was possible to design baits to 205 031 of the ECRs or 99% of the GENCODE exome target regions (Table 1). The total size of these final bait regions is 47.9 Mb.
Table 1. Comparison of the coverage of the design target between the three exome sets.
| Nimblegen CCDSa | Agilent CCDSb | GENCODE exome | |
|---|---|---|---|
| No. of bait regions | 197 218 | 316 000 | 406 539 |
| Genome coverage (Mb) | 34.1 | 37.6 | 47.9c (35.2)d |
| ECRs covered (%) | 150529 (72.7) | 164225 (79.3) | 205031 (99.0) |
| Transcripts covered (%) | 66828 (81.0) | 71279 (86.4) | 81204 (98.4) |
| Genes covered (%) | 28203 (76.5) | 30030 (81.5) | 35989 (97.7) |
NimbleGen Sequence Capture 2.1M Human Exome Array.
Agilent SureSelect Human All Exon Kit.
Total length of bait regions including flanking regions.
Theoretical length without flanking regions.
Sequence capture and sequencing
In all, 15 μg of DNA diluted in TE was sheared to 100–400 bp using a Covaris S2 (Covaris, Woburn, MA, USA). Sheared samples were quantified on a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA), and 7.5 μl of COT 1 DNA at 100 ng/μl was added. This library was lyophilised in a vacuum concentrator to a pellet and resuspended in 3.4 μl of ultrapure water. Following Agilent's SureSelect protocol,14 10 μg of sheared DNA were end repaired and polyA tailed, and Illumina-sequencing adapters were ligated to the resulting fragments using the Illumina (San Diego, CA, USA) Paired-End DNA Sample-Prep protocol, except that the gel-size selection step was replaced with a purification using magnetic bead-based solid phase reversible immobilisation (SPRI) beads (following Agilent's protocol14). The capture library was prepared by mixing 5 μl of the oligo capture library, 1.5 μl of ultrapure water and 1 μl of 1:1 dilution of RNase block. In all, 500 ng of each sample library was hybridised to the appropriate bait set in PCR plates on a thermocycler at 65 °C for 24 h (following the manufacturer's protocol with the modification that no prehybridisation PCR was performed). The capture was performed according to the manufacturer's protocol with streptavidin-coated Dynal beads (Invitrogen, Paisley, UK), and captured samples were washed three times using SureSelect wash buffers with a series of incubation steps. The samples were cleaned up using Mini Elute columns (Qiagen, Hilden, Germany) and eluted in 50 μl of PCR-grade water. Eluted samples were amplified using a master-mix containing 2 m MgCl2, 0.2 m dNTPs, 0.5 μ PE.1, 0.5 μ PE.2 and 3 units of Platinum Pfx DNA Polymerase (Invitrogen) per sample. Samples were aliquoted into three individual wells of a plate and amplified using the following conditions: 94°C for 5 min, followed by 20 cycles of 94°C for 15 s, 58°C for 30 s, 72°C for 30 s and a final extension of 72°C for 5 min. PCR products were purified using SPRI beads before sequencing. All data represent results of one capture reaction for each sample. Captured libraries were sequenced on the Illumina Genome Analyzer 2 platform as paired-end 54-bp reads according to the manufacturer's protocol. The presequencing preparation time is about 3 days, where sonication and library creation take ∼1 day, and hybridisation and amplification 1 day each.
Bioinformatics analysis
Reads were aligned to the human genome (NCBI36) using the MAQ software package v0.7.1.15 Base qualities were recalibrated using the Genome Analysis Toolkit v1.0.3540 (http://www.broadinstitute.org/gsa/wiki/index.php/The_Genome_Analysis_Toolkit) and duplicate fragments marked using Picard v1.17 (http://picard.sourceforge.net/). SNPs were called using SAMtools v0.1.716 and GATK, and the intersection of the resulting call sets in the target regions (39.3 Mb) with a sequence read depth of ≥8 × were reported. Coverage comparisons of the different target set locations were done using BEDTools v.2.6.0.17
Results
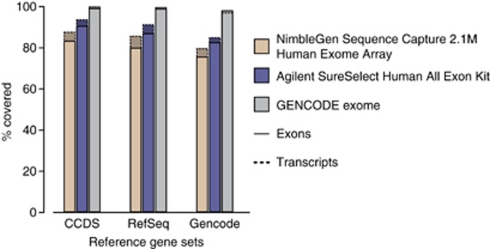
A comparison of the coverage of the bait/oligonucleotide positions of the available CCDS-based exome sets and the GENCODE exome with the set of GENCODE design targets (Table 1) illustrates the increased coverage of our extended target set. The bait positions of the GENCODE exome cover 99% of the targets, which represents an additional 59 600 exons available for capture that are not present in either one of the CCDS-based sets (Supplementary Data 3). The missing 1% consists of regions where reliable bait design was not possible. Comparison of exon and transcript coverage between bait/oligonucleotide locations of the available exome sets and the GENCODE exome, and three current reference gene sets (Figure 1), shows that in all cases, the GENCODE exome covers a greater percentage of the reference gene sets. For example, there is an additional 9% of the exons from the CCDS database and 12% of the exons from RefSeq covered by our expanded target set.
Figure 1.
Comparison of exon and transcript coverage between oligonucleotide locations of the available exome kits and current reference gene sets (CCDS database March 2010, RefSeq genes March 2010 and GENCODE version 3c). The histogram shows the near-complete coverage by the GENCODE exome of all reference sets. Full data are given in Supplementary Table 1.
The content present in the GENCODE exome exclusively consists of 38 933 cluster regions, which contain 5594 additional genes of the design target. The 4363 distinct Ensembl-53-based genes of this set contain 1881 (43%) genes that have an official HGNC identifier, 711 (16%) that are linked to an OMIM entry and 1410 (32%) that have Gene Ontology annotation (Supplementary Data 4). In all, 41% (1809) of these genes have no external annotation of this kind and as such represent novel genes, which could prove to be an important source of variation. The content of repetitive/low-complexity sequence in the bait sets is comparable. The ratios of bases masked by RepeatMasker, Dust and TRF against the total bases in the sets are Nimblegen CCDS: 0.027, Agilent CCDS: 0.021 and GENCODE exome: 0.027 (Supplementary Table 3). A comparison with a sequence uniqueness mask is given in Supplementary Table 4 and supports these findings. The list of 5594 genes and regions targeted by the GENCODE exome exclusively is available as supplementary data and on our ftp site (http://ftp.sanger.ac.uk/gencode/exome), as well as data for the full GENCODE exome and the initial design target. The 406 539 bait locations are supplied as a Distributed Annotation System data source as well (das.sanger.ac.uk/das/Exome), which can be displayed in genome browsers like Ensembl (version 53; http://tinyurl.com/browse-exome).
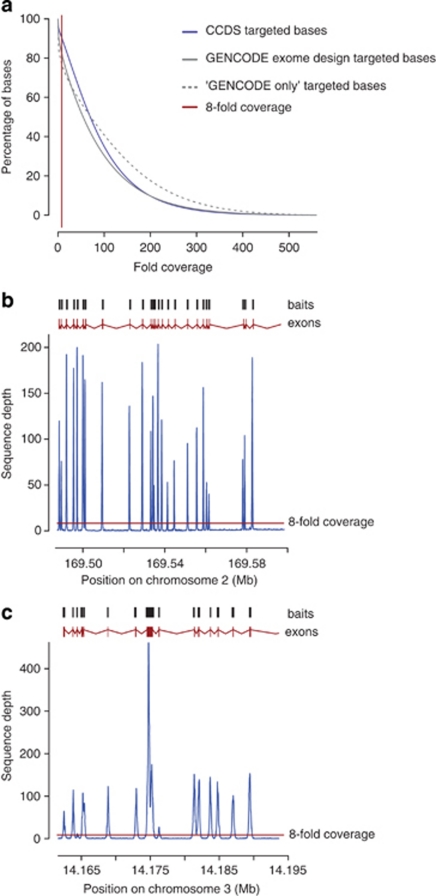
To evaluate the performance of the GENCODE exome, DNA from three HapMap individuals (NA12878, NA07000 and NA19240) was subjected to sequence capture using both the Agilent SureSelect Human All Exon kit and baits designed to the GENCODE exome. In addition, to evaluate the performance using DNA from clinical samples, DNA from seven individuals recruited from a clinical neurological unit was subjected to sequence capture using baits designed to the GENCODE exome. All samples were sequenced as described in the methods section. On average, 97% of reads could be successfully mapped back for both the GENCODE and the Agilent CCDS set. Full details of the sequence yield and reads mapping back to target are given in Supplementary Table 2 (coverage was reported only using reads with a mapping quality of ≥10). The average fold coverage for the HapMap exomes for the CCDS-based targets was 73-fold from 9.2 Gb of sequence and for the GENCODE exome, 82-fold from 11.5 Gb of sequence. The average fold coverage for the clinical samples was 58-fold from 7.5 Gb of sequence. On average for the HapMap samples, 96% of targeted bases were covered at least once and 90% were covered at greater than or equal to eightfold for the CCDS exome, with similar figures for the GENCODE exome of 92 and 83% (Figure 2a). The clinical samples gave an average for the GENCODE exome of 95% of targeted bases covered at least once and 88% covered at greater than or equal to eightfold (Supplementary Figure 1). The results demonstrate that on average, the GENCODE-only regions perform equally to the CCDS regions.
Figure 2.
Coverage achieved by the GENCODE exome. (a) Cumulative fold coverage plot for HapMap samples captured with Agilent SureSelect Human All Exon Kit (CCDS), the GENCODE exome, and the regions covered by the GENCODE exome only. Similar data are presented for the clinical samples and the GENCODE exome only in Supplementary Figure 1. In all cases, the thin red vertical line indicates a fold coverage of eightfold, the preferred coverage required for variant calling. (b and c) Detailed view of the average sequence depth of the seven clinical samples across the entire gene region post sequence capture of two example genes that are unique to the GENCODE exome: (b) ABCB11 and (c) XPC. In the upper part of each panel, the positions of the baits from the GENCODE exome are given as dark grey boxes above the exon structure (adapted from the Ensembl genome browser) of each gene in red. The increased sequencing depth of the eights exon of XPC is caused by good coverage of this larger exon with eight different bait sequences, whereas the other smaller exons are covered by one or two baits.
An average of 22 271 SNPs, of which 2.6% were novel, were found for the HapMap GENCODE exomes compared with 18 554, of which 1.7% were novel, for the CCDS-based exome (Table 2; it should be noted that for most samples, only one lane of the sequencing machine was used. Thus, the sequencing depth does not allow to identify all possible variants, slightly underestimating the number of identified variants). In this instance, novel is defined as not being present in dbSNP18 (version 130) or 1000 Genomes project (1000 Genomes Project Consortium, http://www.1000genomes.org, released on 26 March 2010). An average of 21 866 variants, of which 4.2% were novel, was found in the clinical samples. The clinical samples had been previously genotyped on the Illumina 660 K chip that allowed the concordance rate of the variants found in common with exome sequencing using the GENCODE exome to be calculated at 99.8%. Of the 62 sites, which were discrepant between array genotyping and sequencing, 47 were discrepant only in one sample, suggesting that the number of systematic genotyping errors is low. The ratio of STOP codons gained is approximately in proportion to the size of the exome being captured, suggesting that the extra material in the GENCODE exome does not represent or select for a significant excess of pseudogenes (1.2:1 for the CCDS-based exome in comparison with 1.8:1 for the GENCODE exome). The 22 002 SNPs found on average in the GENCODE exome-captured samples included a mean per sample of 9006 non-synonymous variants, 9424 synonymous variants and 91 stop-gained variants. Therefore, on average, 268 synonymous variants, 256 non-synonymous variants and 2.6 stop-gained variants were found per megabase of the 35.2-Mb targeted genomic sequence, corresponding to a total of 626.6 variants/Mb. In the CCDS-based exome-captured sample among the 18 554 coding SNPs found on average, there was a mean per sample of 7585 non-synonymous variants, 8880 synonymous variants and 45 stop-gained variants, corresponding to 512 variants/Mb.
Table 2. SNP-calling results from clinical and HapMap samples using GENCODE and Agilent CCDS exome captures.
| Sample name | Sanger no. 1 | Sanger no. 2 | Sanger no. 3 | Sanger no. 4 | Sanger no. 5 | Sanger no. 6 | Sanger no. 7 | NA12878 | NA07000 | NA19240 | NA12878 | NA07000 | NA19240 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bait library | GENCODE exome | Agilent CCDS | |||||||||||
| SNPs (polymorphic sites only) | 21 170 | 21 529 | 21 052 | 21 445 | 21 124 | 23 612 | 23 276 | 20 780 | 21 513 | 24 520 | 16 732 | 17 014 | 21 915 |
| dbSNP, % (version 130) | 93.7 | 93.5 | 93.7 | 93.8 | 92.1 | 93.5 | 93.5 | 96.5 | 94.1 | 94.8 | 98.0 | 95.2 | 95.5 |
| dbSNP/1000G, % (26/03/10 pilot 1) | 96.3 | 95.1 | 96.3 | 96.1 | 94.7 | 96.1 | 96.1 | 97.7 | 97.2 | 97.3 | 99.0 | 97.9 | 98.1 |
| Hets | 12 604 | 13 241 | 12 938 | 13 153 | 13 297 | 14 476 | 14 321 | 12 675 | 13 588 | 16 121 | 10 094 | 10 583 | 14 414 |
| Ti/Tv | 3.029 | 2.996 | 3.036 | 3.025 | 2.930 | 3.021 | 3.120 | 3.069 | 3.112 | 3.138 | 3.235 | 3.258 | 3.322 |
| Concordanta | 4638/4648 | 4706/4716 | 4649/4654 | 4569/4582 | 4639/4652 | 5204/5213 | b | 10 491/10 564 | 10 862/10 904 | 10 720/10 810 | 8850/8909 | 9057/9088 | 9795/9877 |
| Concordant % | 99.78 | 99.79 | 99.89 | 99.72 | 99.72 | 99.83 | b | 99.31 | 99.61 | 99.16 | 99.34 | 99.66 | 99.17 |
| Synonymous | 9196 | 9249 | 9072 | 9191 | 8948 | 10 220 | 10 111 | 8480 | 9207 | 10 568 | 7979 | 8133 | 10 528 |
| Synonymous/Mb (35.2 Mb) | 261.25 | 262.76 | 257.73 | 261.11 | 254.21 | 290.35 | 287.25 | 240.91 | 261.57 | 300.23 | 226.68 | 231.05 | 299.10 |
| Non-synonymous | 8608 | 8804 | 8692 | 8828 | 8696 | 9634 | 9385 | 8758 | 8703 | 9958 | 6863 | 6976 | 8918 |
| Non-synonymous/ Mb (35.2 Mb) | 244.55 | 250.12 | 246.94 | 250.80 | 247.05 | 273.70 | 266.62 | 248.81 | 247.25 | 282.90 | 194.97 | 198.18 | 253.36 |
| Stop gained | 86 | 85 | 80 | 89 | 128 | 87 | 95 | 80 | 83 | 99 | 44 | 40 | 51 |
| Stop gained/Mb (35.2 Mb) | 2.44 | 2.41 | 2.27 | 2.53 | 3.64 | 2.47 | 2.70 | 2.27 | 2.36 | 2.81 | 1.25 | 1.14 | 1.45 |
| SNPs (polymorphic sites only) – within GENCODE-only ECRsc | 5179 | 5212 | 5117 | 5162 | 5162 | 5414 | 5424 | 5017 | 5319 | 5887 | |||
| SNPs (polymorphic sites only) – within GENCODE-only ECRs/Mbc | 691.77 | 696.17 | 683.48 | 689.50 | 689.50 | 723.16 | 724.49 | 670.13 | 710.47 | 786.33 | |||
Concordant SNPs were compared with Illumina 660K chip GenCall genotypes for clinical samples or HapMap3 genotypes for HapMap samples NA12878, NA07000 and NA19240.
Missing data.
Excluding flanking regions.
Discussion
The GENCODE gene set as the basis for our exome design provides a more complete set of targets, as it is a merger of the slower but thorough manual Havana and the genome-wide automatic Ensembl annotation. Both Havana and Ensembl are part of the CCDS consortium, and all of the agreed CCDS structures at the time of construction have been incorporated into the new target set.
The new design includes relevant genes for disease-associated variant and mutation discovery. Among the genes in the new, expanded set are members of well-characterised gene families, which are associated with important medical conditions. For example, 43 genes are linked to ion channel activity. Mutations in ion channel genes are known to cause a range of channelopathies, including arrhythmias and inherited paroxysmal neurological disorders.19 Also, the recently identified MLL2 gene linked to the Kabuki syndrome20 was not covered in the CCDS exome but is now represented in the expanded GENCODE exome. There are 70 genes linked to kinase activity. Deregulated protein kinase activity is frequently associated with, and members of the protein kinase family are commonly mutated in, cancer and are thus desired to be covered in cancer sequencing studies. Furthermore, protein kinases are considered to be the targets for the development of new anticancer therapies.21
The sequence capture of the clinical samples for two genes that are targeted by the GENCODE exome only, ABCB11 and XPC, (Figures 2b and c) demonstrates that we have been able to design baits for genes that are unique to the GENCODE exome, and capture them efficiently, with a high degree of specificity. Each exon is covered by a read depth of substantially more than eightfold, which is preferred for variant calling. ABCB11 and XPC genes are already associated with disease, and provide examples of candidate disease genes that are missing from the existing exome-sequencing kits. Indeed, use of the GENCODE exome has already allowed the identification of a pathogenic mutation in a gene causing an autosomal recessive Dwarfism syndrome, which would not have been discovered using a standard CCDS-based exome.22
The advent of the GENCODE exome represents a substantial improvement to the currently available designs for exome sequencing, allowing the capture of a more complete target. We estimate that we were able to call variants in 84% of the total GENCODE exome. This fraction of callable exome regions is likely to increase further with improving sequencing technology and sequencing depths. The GENCODE exome design is being used by the International Cancer Genome Consortium (ICGC; http://www.icgc.org) for their exome-sequencing programme as part of their aim to obtain a comprehensive description of different tumour types and by the UK10K project (http://www.uk10k.org).
Acknowledgments
We thank Hazel Arbery and Matt Humphries for their work on sequence capture, the sequencing teams at the WTSI, especially Mike Quail and John Burton, as well as Barbara Novak, Carlos Pabon-Pena and Angelica Giuffre for probe design, formulation and validation. This work was supported by the Wellcome Trust (grant numbers WT089062, WT062023 and WT077198). FK is supported by the National Institute of Health (grant number 5U54HG004555). MSC is supported by the National Institute for Health Research Cambridge Biomedical Research Centre. PP is supported by the ECOGENE project (EC grant number 205419), by the EU through the Estonian Centre of Excellence in Genomics and by the Estonian Ministry of Education and Research (grant number SF0180026s09). AP and A-EL acknowledge support from the Academy of Finland (200923). Sequencing data have been deposited at the European Genome-Phenome Archive (http://www.ebi.ac.uk/ega/) under accession number EGAS00001000016 and the European Nucleotide Archive (http://www.ebi.ac.uk/ena) under accession ERP000523.
EML is employed by the company selling a product based on the GENCODE exome. The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Scholl UI, Ji W, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Albert TJ, Molla MN, Muzny DM, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, et al. Genome-wide in situ exon capture for selective resequencing. Nat Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Harrow J, Harte RA, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Denoeud F, Frankish A, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7 (Suppl 1:S4.1–S4.9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Aken BL, Ballester B, et al. Ensembl's 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agilent SureSelect protocol http://www.chem.agilent.com/en-US/Search/Library/_layouts/Agilent/PublicationSummary.aspx?whid=60197&liid=5561 .
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübner CA, Jentsch TJ. Ion channel diseases. Hum Mol Genet. 2002;11:2435–2445. doi: 10.1093/hmg/11.20.2435. [DOI] [PubMed] [Google Scholar]
- Ng SB, Bigham AW, Buckingham KJ, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalay E, Yigit G, Aslan Y, et al. CEP152 is a novel genome-maintenance protein and its disruption causes genomic instability in Seckel syndrome. Nature Genetics. 2011;43:23–26. doi: 10.1038/ng.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.