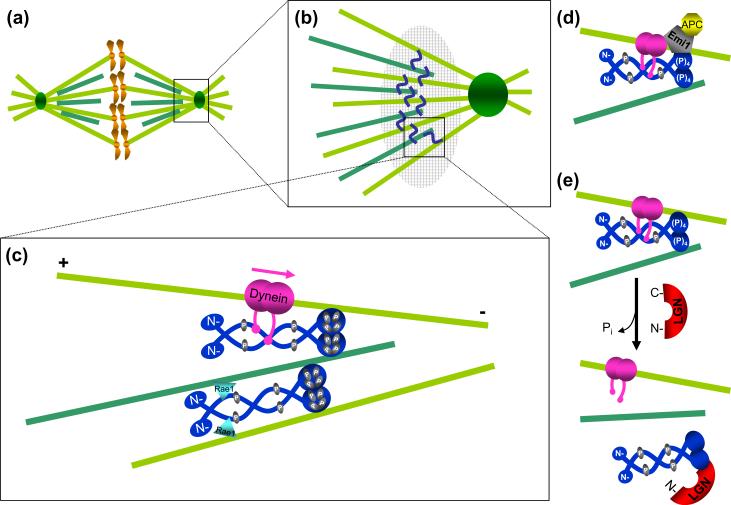
Figure 2.
NuMA functions at spindle poles to bundle and tether microtubules. (a) Schematic of the mitotic spindle, representing kinetochore (light green) and non-kinetochore (dark green) microtubules. The spindle pole (square box) is enlarged in (b). NuMA (blue squiggle) cross-links kinetochore and non-kinetochore microtubules at the spindle pole. NuMA containing complexes involved in microtubule linkage at the spindle poles form a structure equivalent to a “spindle pole matrix” (large hatched oval). (c) Proteins interacting with NuMA at spindle poles (enlarged square box from (b)). During nuclear breakdown, NuMA is phosphorylated (yellow) at four putative p34cdc2 sites (threonine 2000, 2040, 2091, serine 2072) [23, 25]. Mitotically phosphorylated NuMA associates with dynein (pink), which carries it in a minus-end directed fashion (pink arrow) and deposits it at spindle poles where it functions in spindle maintenance via microtubule tethering [20]. A NuMA dimer (dark blue, top) cross-links microtubules through interactions mediated by its C-terminal microtubule binding domain and the associated dynein (pink) [6, 7]. Rae1, a messenger RNA transport protein (blue triangle), may also mediate binding of the NuMA N-termini to microtubules [38], thus transiently generating a higher level of parallel microtubule bundling (bottom microtubules). (d) Emi1 (gray), an inhibitor of Cdc20 dependent activation of the anaphase promoting complex (APC), has been proposed to exist in a complex with dynein and NuMA prior to anaphase [28], as a means of sequestering APCCdc20 (yellow) away from the general cytoplasm. (e) A possible mechanism for NuMA dissociation from microtubules. LGN (red), by competing with microtubules for NuMA binding, acts as a negative regulator of NuMA bundling [7] and may be responsible for the relocalization of a NuMA subset from the spindle pole to the cell cortex.