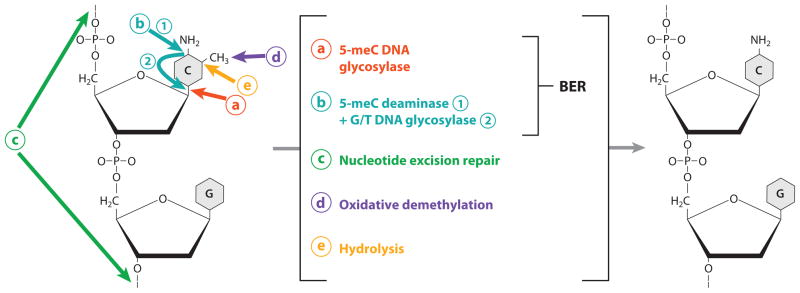
Figure 2.
Diagram showing various possible levels and mechanisms of active DNA demethylation. (a) Base excision repair (BER) initiated by 5-methylcytosine (5-meC) DNA glycosylase. This is the predominant mechanism in plants but may also function in mammals. (b) Base excision repair initiated by coupled activities of 5-meC deaminase that converts 5-meC to T, and G/T mismatch DNA glycosylase that corrects the G/T mismatch. This appears to be the predominant mechanism in mammals but may also play a role in plants. (c) Nucleotide excision repair that removes methylated CpG dinucleotides. (d ) Oxidative removal of the methyl group. (e) Hydrolytic removal of the methyl group, releasing it as methanol.