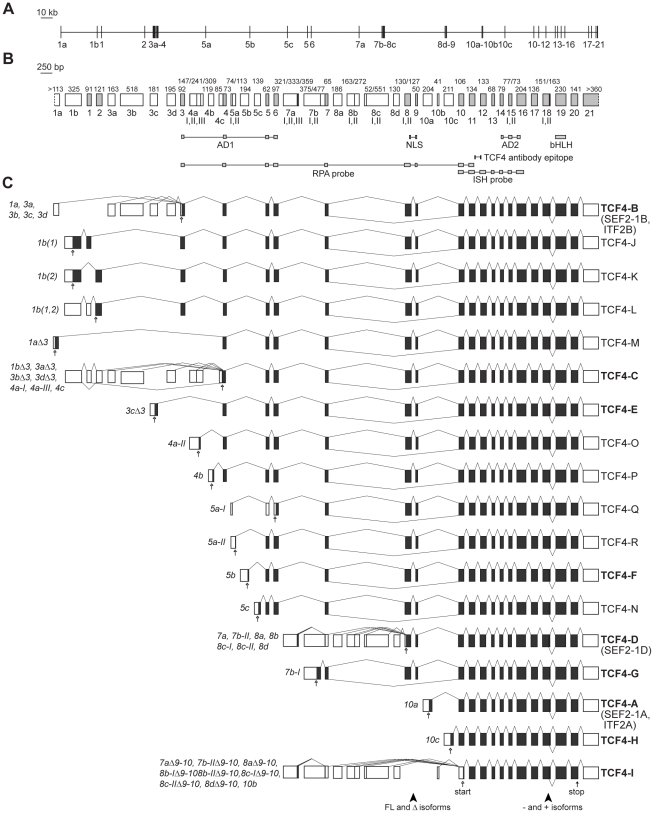
Figure 1. Structure and alternative splicing of the human TCF4 gene.
TCF4 genomic organization with (A) introns drawn in scale or (B) exons drawn in scale. White boxes mark 5′ exons and light grey boxes represent internal or 3′ exons. Exon names are shown below the boxes. Roman numerals designate alternative splice donor or acceptor sites. Numbers above the exons indicate their sizes in bps. The regions encoding the respective domains of TCF4, the NLS identified in this study and the epitope of the used TCF4 antibody are indicated below the gene structure. Locations of RPA and ISH riboprobes used in this study are also shown. AD, transcription activation domain; bHLH, basic helix-loop-helix domain; NLS, nuclear localization signal; RPA, ribonuclease protection assay; ISH, in situ hybridization. (C) TCF4 alternative transcripts grouped together according to the encoded TCF4 protein isoform. Translated and untranslated regions are indicated as dark grey and white boxes, respectively. Transcripts are designated with the name of the 5′ exon and, if needed, with the number of the splice site used in the 5′ exon. Excluded internal exons are shown with the symbol Δ and included internal exons in parentheses, if necessary. The names of the protein isoforms are shown at the right. The isoforms cloned in this study are brought in bold. The position of the first in-frame start codon for each transcript and stop codon are shown with empty and filled arrows, respectively. Arrowheads at the bottom of the panel point to the regions of alternative splicing giving rise to full-length (FL) and Δ, − and + isoforms.