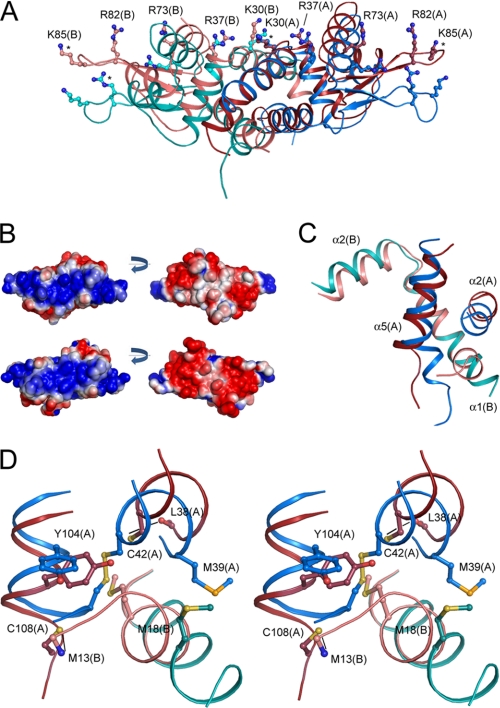
FIGURE 3.
The structural basis for the redox-regulated DNA binding in BigR. A, basic residues potentially involved in DNA interaction in oxidized (blue) and reduced BigR (red) are represented in ball-and-sticks format. Dark and light colors are used to distinguish monomers A and B, respectively. Residues marked with an asterisk are replaced by alanines in the refined structure, and their side chains were modeled to produce this image. B, electrostatic surface of reduced (top) and oxidized (bottom) BigR showing differences in the basic DNA-binding region as well as in the negatively charged surface of the opposite face of the dimers. The bonds for potential contour map visualization are ± 2 kT/e. C, comparison of the secondary structure elements in oxidized (blue) and reduced (red) BigR showing the N terminus of helix 1 in between helices 2 and 5 in the reduced structure. Dark and light colors correspond to monomers A and B, respectively. D, stereo view of the Cys-42 and Cys-108 neighborhood depicting a network of sulfur-containing residues.