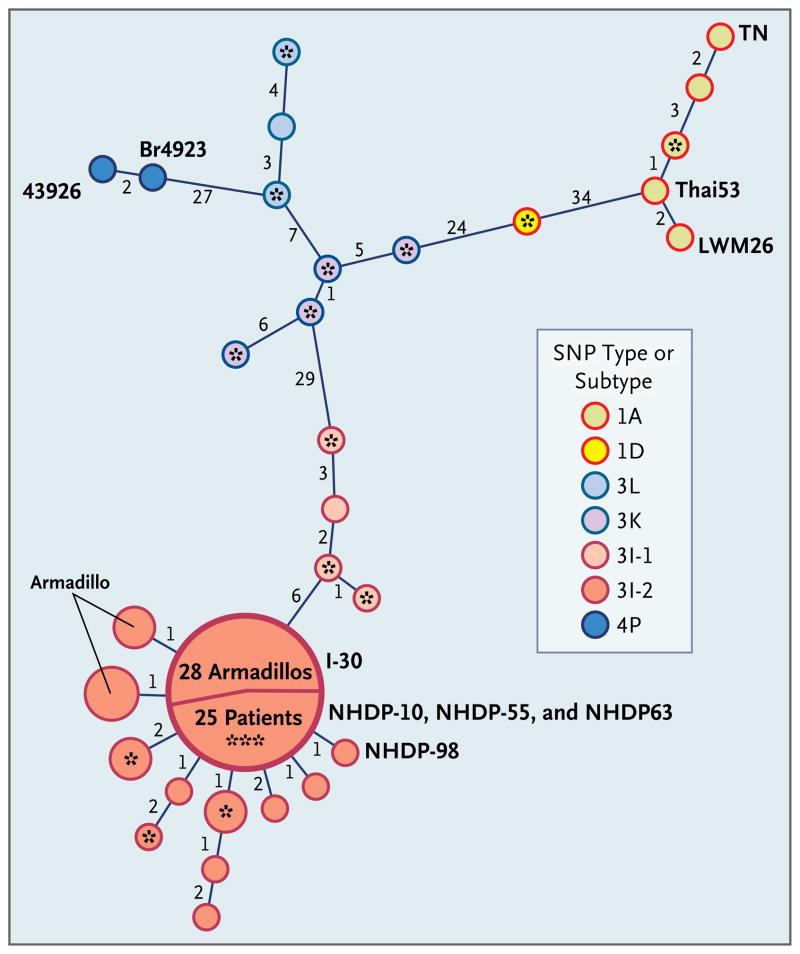
Figure 3. Minimum-Spanning Phylogenetic Tree of Mycobacterium leprae Genotypes Based on Analysis of Single-Nucleotide Polymorphisms (SNPs) and Variable-Number Tandem Repeats (VNTRs).
Minimum-spanning-tree analysis was performed with the use of combined VNTR and SNP data from human and armadillo M. leprae strains. Each circle represents a genotype (human unless marked as armadillo) based on the combined data, with the circle size directly proportional to the number of strains with the corresponding genotype. Numbers along the links between circles indicate the number of loci that differ between the genotypes on either side of the link. Three fully sequenced reference M. leprae strains (TN, Thai53, and Br492322,29) are labeled, as are two other reference strains (LWM26 and 43926) of foreign origin. Samples from patients with a history of foreign residence are indicated with an asterisk (with three asterisks indicating three patients). The 114 polymorphisms investigated include 84 SNPs described previously22 and 30 identified during our study; 10 VNTRs were also analyzed. The large circle illustrates the predominance of the 3I-2-v1 M. leprae genotype in our study, with 25 patients and 28 armadillos having this identical genotype.