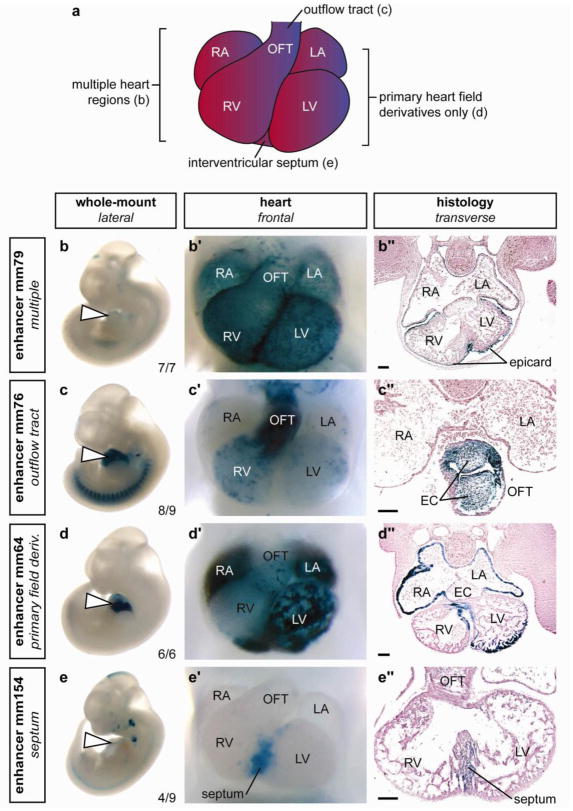
Figure 3. Examples of the diverse structural and cell type specificities of p300 ChIP-seq identified cardiac enhancers.
a) Schematic of mouse embryonic day 11.5 heart. b–e) Side views of whole embryos, b′–e′) magnified ventral views of hearts and b″– e″) transverse sections through the heart region are shown for each of four representative X-gal-stained embryos with in vivo enhancer activity at e11.5. Element ID and reproducibility of expression patterns are indicated alongside whole embryo images. b) Enhancer with activity exclusively in epicardium in all anatomical regions of the heart. c) Enhancer with activity primarily in outflow tract endocardium and in all endocardial cushion (EC) mesenchyme. d) Enhancer primarily active in derivatives of the primary heart field (atrial and ventricular myocardium and a small region of the interventricular septum). e) Enhancer with activity predominantly in the muscular portion of the interventricular septum. RA, right atrium; LA, left atrium; RV, right ventricle; LV, left ventricle; OFT, outflow tract; epicard, epicardium. The bars in panels showing transverse sections are equal to 100 μm. The complete in vivo expression dataset is available online40.