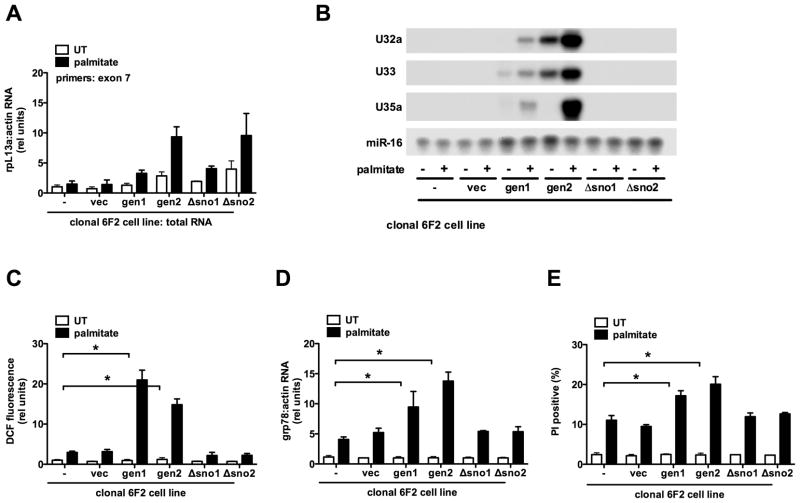
Figure 4. rpL13a genomic sequence containing box C/D snoRNAs complements mutant 6F2 cells.
(A–E) Mutant 6F2 cells were stably transfected with a construct containing genomic rpL13a murine sequence, genomic rpL13a murine sequence with deletion of U32a, U33, U34, and U35a, or the empty vector. Clonal cell lines were isolated and analyzed untreated (UT) or supplemented with palmitate for 48 h. Graphs show untransfected 6F2 cells (−), a stable 6F2 line harboring vector sequences only (vec), and lines expressing genomic (gen1, gen2) or snoRNA-deleted (Δsno1, Δsno2) rpL13a sequences. (A) cDNA synthesis was primed with random hexamers and rpL13a expression analyzed by qRT-PCR and normalized to β-actin. Primers were chosen from exon 7 sequence that is completely conserved between mouse and hamster. (B) Small RNA was used in RNase protection assay with murine-specific rpL13a snoRNA probes or mir-16 probe as control. (C) ROS generation was quantified by DCF labeling and flow cytometry. (D) cDNA synthesis was primed with oligo dT and grp78 expression analyzed by qRT-PCR and normalized to β-actin. (E) Cell death was assessed by PI staining and flow cytometry on 104 cells/sample. All data expressed as mean ± SE for 3 independent experiments. * p < 0.01. (See also supplemental Figure 4.)