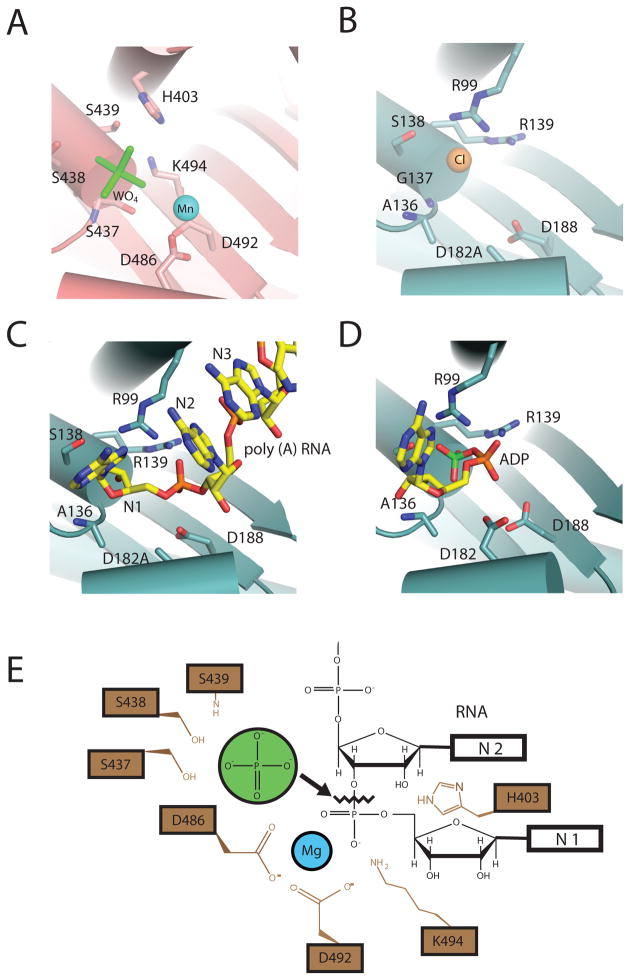
Figure 4. Phosphorolytic exoribonuclease catalytic mechanism for PNPase and the archaeal exosome.
A) PNPase active site. A composite structure is depicted that shows the phosphate-mimic tungstate (green) and magnesium ion-mimic manganese (blue sphere) (PDB ID = 1E3P and 3GME). Residues that bind “phosphate” include: S439, S438, and S437. Residues that coordinate “magnesium” are D486 and D492. B) S. solfataricus exosome active site. The phosphate-mimic chloride is shown as a yellow sphere, and residues that bind the proposed phosphate-mimic chloride are: A136, G137, and S138 (PDB ID = 2BR2). C) D182A mutant S. solfataricus in complex with a five-nucleotide poly(A) RNA substrate (PDB ID = 2C38). Nucleotides are colored yellow and numbered in such a manner that the first nucleotide (N1) is at the 3′OH end. D) S. solfataricus in complex with the product ADP (PDB ID = 2C39). ADP is colored in yellow with the α-phosphate colored orange and β-phosphate colored green. E) Proposed phosphorolytic exoribonuclease mechanism. Ser437 and Ser439 provide a binding pocket for phosphate (green). Asp486 and Asp492 coordinate a magnesium ion. The magnesium, with the aid of K494 and H403, positions the bridging phosphate between N1 and penultimate N2 nucleotides to facilitate in-line attack by the phosphate.