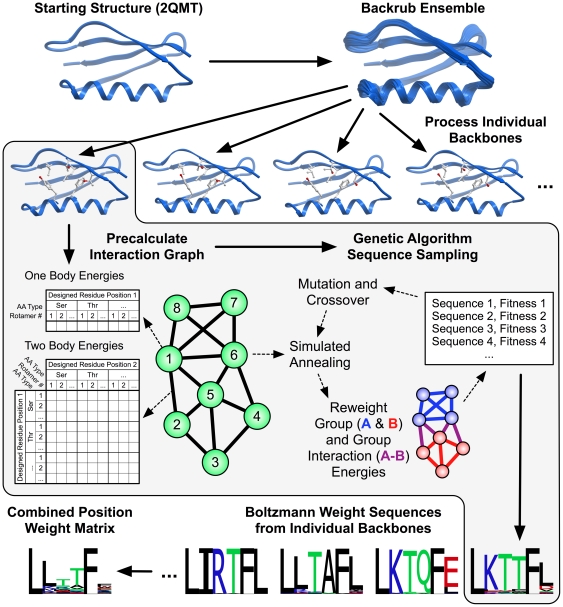
Figure 1. Scheme for predicting the tolerated sequences for a protein fold or interaction.
The input is at least one protein structure from the protein structure databank (2QMT in the example). Rosetta first creates an ensemble of backbone conformations using the backrub method [31], then predicts sequences consistent with each conformation in the ensemble, scoring each trial sequence–structure combination using the Rosetta score12, and finally combines the sequences into a predicted sequence profile. This approach ignores potential covariation between side chains. To speed up calculations, the scoring function is split into one-body terms describing the intrinsic energy of a particular residue conformation, and two-body terms between residues; these residue-residue interaction terms are assumed to be pairwise additive. One- and two-body terms are pre-calculated and stored in an interaction graph [42] such that optimization of sequence–structure combinations for entire proteins only takes seconds using look-up tables of interaction energies. For the interaction graph, vectors of residue self-energies (one body) are stored on the vertices (green circles) and matrices of residue interaction energies (two body) are stored on the edges (thick black lines). Computed interaction energies within proteins, between proteins, or between groups of residues can be reweighted to generate custom fitness functions for specific applications. This flexibility in scoring residue groups allows modeling of separate requirements, such as those to maintain residues required in an interaction interface with a binding partner. Group and group interaction reweighting is typically only done for protein-protein interactions. (For the monomeric GB1 domain shown here, no reweighting was applied.)