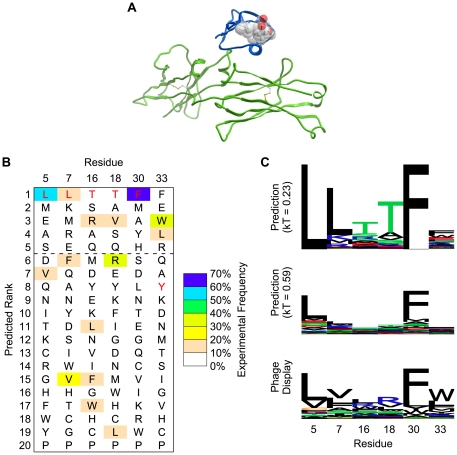
Figure 2. Prediction of tolerated sequences for GB1 fold stability.
Frequently observed amino acids in phage display are enriched in the GB1 prediction. A. The structure (PDB code 1FCC) of Streptococcal GB1 (blue) is shown bound to the Fc domain of human IgG (green). The core and peripheral residues that were randomized in phage display are shown with sticks and transparent spheres. The side chain atoms (starting at C-beta) of these amino acids are at least 7 Å away from any atom of the Fc domain, making residues selected at these positions unlikely to interact directly with the Fc domain. B. Amino acids are ranked individually for each sequence position by computationally predicted frequency (using the Boltzmann factor kT = 0.23, as described in the main text). Wild type residues, which were used in protein ensemble generation, are shown in red. The dashed line indicates a typical cutoff of picking the top 5 amino acid choices at each position. C. Sequence logos (LOLA, University of Toronto) are shown for predictions with two different Boltzmann factors. The relative degree of specificity (in terms of bits of information, y-axis) shows good correspondence between prediction and phage display. Increasing the Boltzmann factor lowers the overall specificity and brings the absolute frequencies closer to phage display.