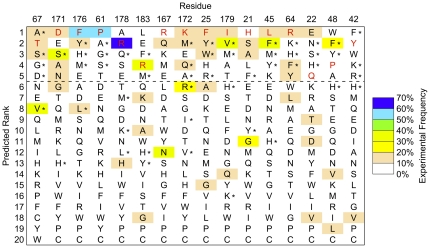
Figure 3. hGH/hGHR interface tolerance prediction.
The generalized Rosetta 3 protocol described here was applied to rank human growth hormone (hGH) amino acids by computationally predicted frequency. The residue positions shown and their ordering are taken from previously published results using the Rosetta 2 protocol (Humphris & Kortemme, Table 2 [23]). Wild type residues, which were used in protein ensemble generation, are shown in red. For each position, an average of 59% of the amino acids observed in phage display (≥10% experimental frequency) are predicted within the top five computationally ranked amino acids (above dashed line). Overall performance was similar to previous results of the Rosetta 2 protocol. Amino acids (other than wild-type) included in the computationally selected library from the Rosetta 2 protocol are indicated with a star. If the same number of amino acids at each position is used as defined in the computational library in [23], Table 2, the Rosetta 3 protocol misses two frequently observed amino acids included by Rosetta 2 (V67 and L176). Conversely, the Rosetta 2 protocol misses three frequently observed amino acids included by Rosetta 3 (S21, A21, and E22). Both protocols share similar false positive predictions. However, the Rosetta 3 histidine reference energy reweighting (see Methods) eliminates 6 out of 8 histidine false positives (H*).