Figure 1. Tumors and immunosuppressive stimuli upregulate GILZ expression in DCs while activation decreases it.

(A) To determine whether the immunosuppressive environment of tumors would upregulate GILZ expression in DCs, mice were injected with 1 × 106 A20-HA B cell lymphoma cells. Ten days later, tumor-containing spleens were harvested, single cell suspensions prepared, stained with CD11c, and FACS sorted into a CD11c+ population. RNA was prepared from the DCs from the tumor site, reverse transcribed into cDNA and subjected to qPCR. Shown are the relative values of GILZ expression, normalized to actin, with naïve DCs set to control values of 100%. Minimum of three experiments were combined for each subset of figure 1; statistics shown are paired t tests. For each panel, asterisks denote differences that are significantly different (p<.05) from control.
(B) Tumors upregulated GILZ in DCs via a soluble factor. To assess whether cell-cell contact was required between tumor cells and DCs, and to assess whether other tumor types would increase expression of GILZ, the supernatants from either A20 or B16 melanoma tumor cells were harvested 48 hours after medium change, filtered, and added to cultures of DCs. Twenty four hours later, DCs were harvested and RNA and cDNA prepared for the qPCR assay as in 1A. A paired t test shows significant differences for both tumor types compared to controls.
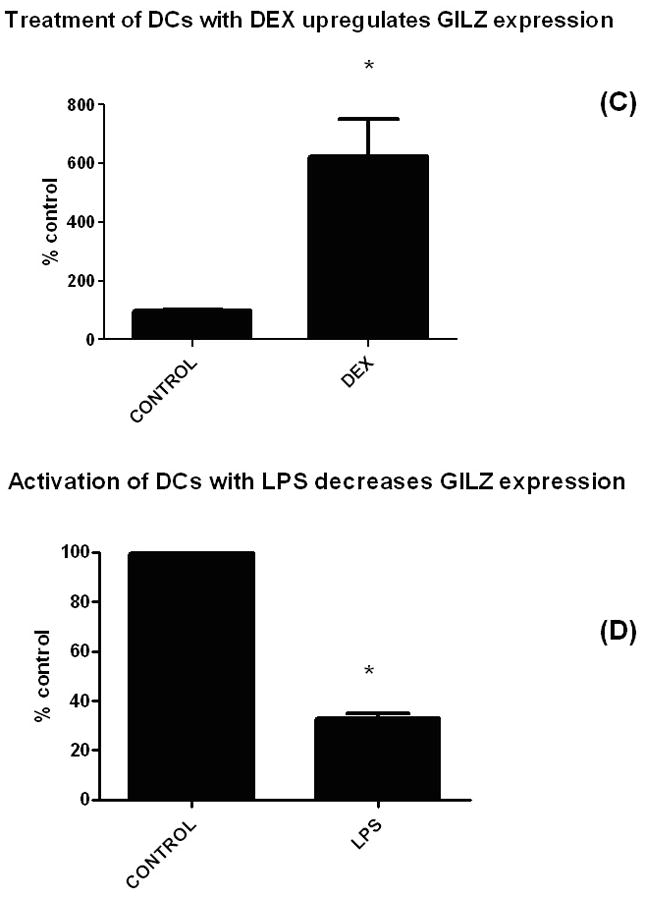
(C) Dexamethasone upregulated GILZ in BMDCs and (D) LPS decreased GILZ expression. BMDCs were exposed to a suppressive (DEX) and an activating (LPS) agent and expression levels of GILZ were measured by qPCR (normalized to expression of actin). For the cultures, the following concentrations were used: DEX treatment (100 nM, overnight); LPS (100 ng/ml, overnight).
