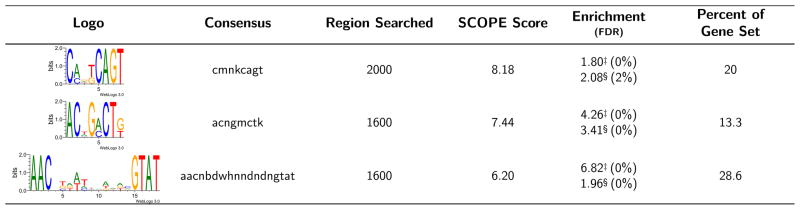
Figure 1.
Sequence Logo indicates predominant nucleotides in the consensus for each motif identified (http://weblogo.threeplusone.com/create.cgi). ‘Percent of Gene Set’ indicates the percent of all 5′ genomic regions bearing the motif of interest. ‘SCOPE Score’ indicates the negative log of the expectation as calculated by SCOPE. “Enrichment” presents the hypergeometric enrichment values (−log10(hypergeometric p-value)) for position weight matrix thresholds of 75% (‡) and 90% (§) of the maximum score with FDRs listed in parentheses. “Region Searched” indicates the 5′ genomic region of interest that yielded the given motif.