Abstract
Like all retroviruses, the Human Immunodeficiency Virus (HIV) selectively packages two copies of its unspliced RNA genome, both of which are utilized for strand-transfer mediated recombination during reverse transcription – a process that enables rapid evolution under environmental and chemotherapeutic pressures. The viral RNA appears to be selected for packaging as a dimer, and there is evidence that dimerization and packaging are mechanistically coupled. Both processes are mediated by interactions between the nucleocapsid (NC) domains of a small number of assembling viral Gag polyproteins and RNA elements within the 5′-untranslated region (5′-UTR) of the genome. A number of secondary structures have been predicted for regions of the genome that are responsible for packaging, and high-resolution structures have been determined for a few small RNA fragments and protein-RNA complexes. However, major questions remain open regarding the RNA structures, and potentially the structural changes, that are responsible for dimeric genome selection. Here we review efforts that have been made to identify the molecular determinants and mechanism of HIV-1 genome packaging.
Keywords: retrovirus, HIV, genome packaging, RNA structure, NMR spectroscopy
Introduction
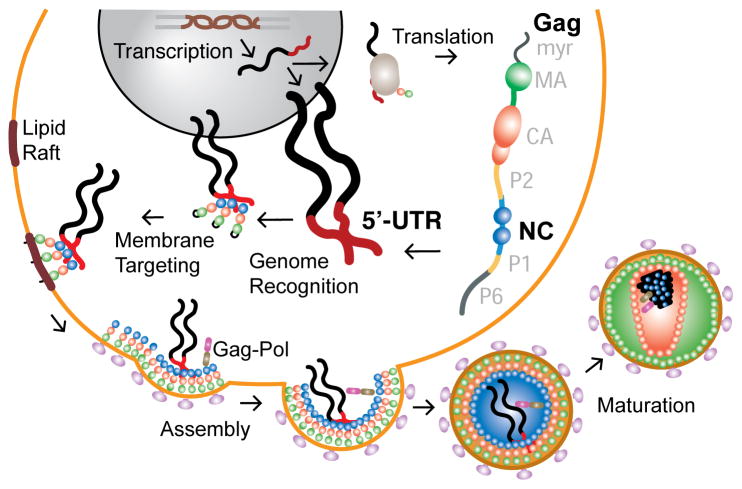
During the Late Phase of the viral replication cycle, the Human Immunodeficiency Virus Type-1 (HIV-1) selectively and efficiently packages two copies of its positive strand, unspliced, 5′-capped and 3′-polyadenylated RNA genome by a mechanism that has been extensively studied (for previous reviews, see1–9) but remains only partially understood. The packaging mechanism efficiently discriminates against the monomeric genome, the spliced viral mRNAs that encode for viral accessory envelope proteins, and the more highly abundant cellular mRNAs2. Packaging is mediated by the retroviral Gag proteins, which can efficiently assemble in the absence of their native genomes by incorporating an equivalent amount of cellular RNAs10–14. Although retroviruses can package essentially any RNA (some mutants even package ribosomes14), RNAs containing the appropriate viral packaging signals are efficiently enriched in assembling virions. The Gag protein contains three independently folded domains (from N- to C-terminus): matrix (MA), capsid (CA) and nucleocapsid (NC), as well as three other unstructured but functionally important segments, Figure 1. Genome selection appears to proceed via the direct binding of NC to conserved RNA packaging signals, called Ψ-sites, that are generally located near the 5′-end of the viral RNA. It now seems likely that a ribonucleoprotein complex comprising a relatively small number of Gag molecules and two copies of the genome is trafficked to plasma membrane assembly sites, where several thousand additional Gag molecules localize and assemble to form an immature virus particle15,16. During or shortly after budding, the Gag proteins are cleaved by the viral protease to produce the mature MA, CA and NC proteins, which rearrange to form the mature and infectious virus particle, Figure 1.
Figure 1.
Cartoon showing selected HIV-1 Late Phase replication events associated with genome packaging and virus assembly. The full-length genome is exported from the nucleus in a Rev-dependent manner and functions as the mRNA for ribosomal synthesis of the Gag and Gag-Pol (by a frameshift; not shown) polyproteins. A small number of Gag proteins (probably a dozen or fewer) form a complex with the dimeric viral genome. It is currently not clear if formation of this initial complex occurs mainly in the nucleus, via transient nuclear import of the Gag protein, or in the cytoplasm (or possibly in both locations). The ribonucleoprotein complex is trafficked to lipid raft assembly sites on the plasma membrane (mediated by the myristylated (myr) MA domain) where further assembly and budding occur.
The requirement for two genome molecules is intriguing since all other viruses contain only a single copy of their genetic material17. Both RNA molecules are utilized for strand transfer-mediated recombination during reverse transcription18,19, but only one DNA allele is generated, and retroviruses are therefore considered “pseudodiploid.” Recombination appears to enhance viral fitness in several ways. It enables strand transfer-mediated read through at sites of RNA damage20,21, which could serve as a mechanism for dealing with what appears to be a relatively fragile genome22, and as a defense against restriction nucleases23. In addition, although cells containing a single integrated provirus can only produce homozygous virions, cells from infected individuals have sometimes been observed to contain several integrated proviruses24–26, which probably explains the very high number of circulating recombinant forms of HIV-127. Thus, strand transfer-mediated recombination from heterozygotes likely serves as a primary pathway for the rapid evolution viruses that are resistant to antiretroviral therapies28. Retroviral genomes exist as weak, non-covalently linked dimers in immature and young virus particles, and the stability of the RNA dimer29–34 increases with virus age, which might be important for subsequent reverse transcription events. The genome also appears to play a structural role in virus assembly, although this function can also be achieved by cellular RNAs13.
As for most other retroviruses, the nucleotides that participate in HIV-1 genome selection appear to reside near the 5′-end of the genome and primarily within the 5′-Untranslated Region (5′-UTR)1–9. Relatively short elements within the 5′-UTR that are independently capable of directing heterologous RNAs into assembling virus-like particles (VLPs) have been identified for some retroviruses (for example, the Rous Sarcoma Virus (RSV)35–40 and Moloney Murine Leukemia Virus (MoMuLV)41,42), but HIV-1 appears to require most of it’s 5′-UTR43–58 as well as downstream nucleotides within the gag coding region59–61 for optimal packaging efficiency. The 5′-UTR is the most conserved region of the HIV-1 genome62,63 (http://www.hiv.lanl.gov/), and in addition to promoting packaging, it also helps regulate or promote transcriptional activation, splicing, primer binding during reverse transcription, and dimerization. For some retroviruses, elements known to be important for packaging reside downstream of the major 5′ splice donor site (SD), providing a potential mechanism for selecting the full length genome and ignoring spliced viral mRNAs64. RNA elements that are believed to facilitate HIV-1 genome packaging reside near elements that promote RNA dimerization3,4,6,8, and since both processes are promoted by the NC domain of Gag, it is likely that genome dimerization and packaging are intimately coupled1,2,65. This now certainly seems to be true for the evolutionarily distant MoMuLV virus66–68. Efforts to elucidate the structural determinants and mechanisms that regulate these activities have been made using a variety of biochemical, in vivo packaging and computational approaches, and there is general consensus that some activities are controlled by well-defined hairpin structures within the HIV-1 5′-UTR48–50,54,57,69–75. However, there is less agreement regarding the structures that regulate genome packaging. Here we review efforts that have been made to understand the mechanism of HIV-1 genome selection, with emphasis on the RNA and protein-structures that appear to play important roles.
Genome Selection is Mediated by the NC Domain of Gag
There is now considerable evidence that genome selection is mediated primarily by the NC domain of the HIV-1 Gag polyprotein. Substitution of the HIV-1 NC domain by that of MoMuLV leads to the preferential packaging of the MoMuLV RNA76 into HIV-1 derived chimeric virions, and conversely, substitution of the MoMuLV NC domain by that of HIV-1 results in preferentially packaging of the HIV-1 genome77. Similar results have been observed for other retroviruses78. Retroviruses belonging to a given genus appear to be more readily capable of packaging each other’s RNAs79–81. Non-reciprocal packaging has also been observed for some combinations of retroviruses82,83, and interestingly, the HIV-1 and mouse mammary tumour viruses (MMTV) preferentially package their native RNAs, even when the NC domains of these retroviruses are swapped84. These findings suggest that the NC domains are not uniquely responsible for RNA selection.
Except for the spumaviruses, all retroviral NC proteins contain one or two copies of a conserved CCHC array motif (Cys-X2-Cys-X4-His-X4-Cys; X = non Cys/His amino acid) that was originally discovered by Henderson and co-workers85 and later proposed by Berg to function as a zinc binding site86. Although early biochemical studies led to suggestions that the arrays have weak affinity for zinc87 and do not contain zinc in virions88,89, subsequent in vivo mutagenesis experiments showed that conservative single atom S to O substitutions in the arrays (Cys to Ser) block both viral replication and genome packaging, consistent with a zinc binding function90. Studies with peptides and both recombinant and virus-derived HIV-1 NC protein confirmed that the arrays are capable of binding zinc with high affinity91–94, and zinc-edge Extended X-ray Absorption Fine Structure Spectroscopy (EXAFS) studies of intact retroviruses eventually provided strong evidence that the arrays are populated with zinc in mature particles95,96. These and other studies44,90,97–99 have provided convincing evidence that the NC zinc knuckles play an essential role in selecting the unspliced genome during retrovirus assembly. Compounds that eject zinc from the NC zinc knuckles have potent antiviral activity in cell cultures100, and several antiviral zinc ejectors have been identified100–103. Unfortunately, poor specificity and toxicity have thus far precluded the therapeutic development of this class of antivirals, although they have been used as reagents in structure probing experiments104.
HIV-1 genome packaging might also be facilitated by the nucleic acid chaperone property of NC, which is known to promote dimerization of viral RNA33,105,106. Recent studies have shown that dimerization of the MoMuLV packaging signal leads to a register shift in base pairs that exposes high affinity NC binding sites, and that the MoMuLV NC protein facilitates the RNA structural changes associated with dimerization66. An important future goal should be to determine if the HIV-1 leader exhibits similar dimerization-dependent NC binding. NC can also stimulate annealing of the appropriate cellular tRNA primer to the primer binding site (PBS), a requirement for the initiation of reverse transcription107,108. In the presence of NC, pausing of reverse transcriptase (RT) is greatly reduced, thus increasing the efficiency of the synthesis of full-length DNA109,110. NC induces annealing of the strong stop cDNA with the complementary region of the 3′-end of the genomic RNA, and increases the strand transfer efficiency during reverse transcription111–115. Mutational analysis indicates that the N-terminal highly basic residues of NC are crucial110, and the two specific zinc-finger architectures also appear to be required for its nucleic acid chaperone activity. As discussed in a recent review by Levin and coworkers116, the mechanism for the chaperone activity likely involves relatively weak and labile NC-RNA interactions that lower the energy barrier for breaking and/or reforming base pairs117–119.
To date, no X-ray crystal structures have been reported for any CCHC zinc knuckle. However, NMR structures have been reported for several isolated zinc knuckle domains120–123, intact NC proteins96,124–129, and NC:RNA complexes66,128,130–137. The zinc knuckles form highly stable mini-globular domains that are structurally distinct from other CCHC-type modules131,138,139. Observation of weak NOEs between aromatic residues of the N- and C-terminal HIV-1 NC zinc knuckles suggested that the domains pack together in solution126, but an alternative model, in which the zinc knuckles are connected by a flexible tether and interact weakly, was proposed on the basis of NMR relaxation and chemical shift analyses140. The knuckles of the HIV-2 and SIV NC proteins appear to interact tightly with each other in the absence of nucleic acids127,128. Tight inter-knuckle packing is clearly observed for HIV-1 NC upon binding to RNAs with high affinity binding sites (see below)132,133,137.
Potential Roles for the MA Domain in Genome Packaging
The MA domain of Gag plays important roles in intracellular trafficking and membrane targeting141–144, and by this means may also be important for genome packaging. In 1993, Arlinghaus and co-workers showed that approximately 18% of the cellular MoMuLV Gag protein is associated with the nucleus, and a role of nuclear Gag in regulating splicing and/or dimerization activities was proposed145. Green and co-workers subsequently reported that mutating two basic residues in the HIV-1 MA domain of Gag can lead to accumulation of Gag and the viral genome in the nucleus146. These mutations also led to a significant reduction in virus particle production, and the particles that were formed were poorly infectious and appeared to contain predominantly monomeric genomes. Earlier studies had shown that the MA domain of Gag contains a nuclear localization signal (NLS)146–150, and these findings suggested that MA also possesses a nuclear export signal (NES)146, and that both signals play a role in the transient shuttling of Gag through the nucleus prior to virus assembly. These findings apparently have not been replicated in other laboratories, which has raised questions about the proposed role of nuclear shuttling by HIV-1 Gag. However, Parent and co-workers reported a similar transient nuclear shuttling activity by the Gag proteins of the Rous sarcoma virus (RSV). In this case, nuclear export could be blocked by specific mutations in MA or treatment agents that inhibit the Chromosome Region Maintenance-1 (CRM-1) dependent nuclear export pathway151,152. Interestingly, a mutation in RSV MA that enhanced membrane binding (Myr1Glu) and blocked nuclear shuttling led to the formation of particles with reduced levels of primarily monomeric genomes153,154. These studies suggested that transient nuclear localization of Gag plays a role in genome packaging. Although arguments have been made that question a nuclear shuttling role for Gag in genome packaging152, Parent and co-workers very recently showed that wild-type levels of RSV genome packaging could be restored by inserting a non-viral NLS into Myr1Glu-Gag154. Thus, although questions remain regarding the ability of HIV-1 Gag to transiently access the nucleus, the more extensive studies with RSV are consistent with a packaging mechanism in which genome recognition occurs in the nucleus, and transient nuclear shuttling of Gag, directed by the MA domain, is required for efficient trafficking of the genome from the nucleus to virus assembly sites on the plasma membrane. Note, however, that very recent studies by Hu and co-workers indicate that dimerization of the HIV-1 genome takes place mainly in the cytoplasm, subsequent to nuclear export155. It is conceivable that different retroviruses use unrelated mechanisms to select their genomes. An alternative (and untested) possibility is that multiple RNA export pathways are available to all retroviruses, and that the dominant pathway either changes temporally with the age of the infected cells or is dependent on conditions employed for the packaging/replication experiments.
The MA domain could also function by interacting directly with the viral RNA. Because they are highly basic, retroviral MA proteins are capable of binding to nucleic acids156–158. Early SELEX experiments identified a short RNA sequence that can bind to MA with high affinity (~10−9 M), but the sequence did not correspond to sequences within the viral genome and the biological relevance of these findings was not clear159. A subsequent SELEX study by a different group identified a different RNA with high affinity for the MA domain of Gag (~5 × 10−7 M) that possessed a sequence similar to one in the Pol gene160. Although mutations in Pol designed to disrupt interactions with RNA led to a delay in replication, the relevance of the specific interactions proposed by these studies remains to be firmly established. More recent studies examined the abilities of nucleic acids and liposomes to compete for the myristylated MA protein and the myr-MA domain of a MA-CA construct. These studies showed that liposomes can only compete efficiently for MA if they contain phosphatidylinositol-4,5-bisphosphate (PIP2), a cellular factor required for Gag membrane targeting161,162, suggesting that MA interactions with the viral genome may contribute to membrane selectivity163. Similar conclusions were reached in a subsequent study164. Other studies have shown that the ability of HIV-1 Gag to promote tRNA binding to a PBS oligonucleotide is stimulated by the binding of PIP2 to the MA domain165. It therefore seems plausible that MA-RNA interactions, mediated by PIP2-dependent membrane binding, could function in the regulation of Gag’s chaperone activity, possibly inhibiting primer binding prior to membrane binding165.
Genomes are Selected for Packaging as Dimers
Early studies by Mangel and co-workers showed that RNAs extracted from the RSV under mild denaturing conditions were dimeric, as determined by sucrose gradient sedimentation, but formed monomers in the presence of the gene-32 protein that unwinds double-helical nucleic acids structures and preferentially binds to single stranded RNAs166. These studies showed that the RNA in virions exists as non-covalently linked dimers, but they did not rule out the possibility that the RNAs might be recognized by Gag as monomers and form dimers during or shortly after assembly. About the same time, Berezesky and co-workers showed that MoMuLV virions produced from cells treated with actinomycin D, which inhibits mRNA synthesis, packaged significantly less viral RNA, and that the RNAs that were packaged were dimeric167. These findings suggested that RNAs might be selected for packaging as dimers prior to virus budding and assembly. MoMuLV particles that were rapidly harvested and mutant virions that lack protease activity, package loosely formed dimers that are readily dissociated by mild heating33, suggesting that protease-induced viral maturation promotes a maturation of the viral RNA to a more stable dimeric form. Similar maturation-dependent RNA dimer stability has been observed for HIV-1105. More recent studies by Laughrea and co-workers revealed that small amounts of monomeric genome could be isolated from very young virus particles (i.e. less than 30min old), and that the monomeric RNAs convert to dimers in a protease- and time-dependent manner168. Sakuragi and co-workers showed that HIV-1 derived RNAs containing an additional, downstream 5′-UTR element (which lacked the PolyA and SD residues, to prevent polyadenylation and aberrant splicing) could be efficiently packaged into HIV-1 virus-like particles as monomers, suggesting that it is the structure of the dimeric 5′-UTR that is responsible for packaging169. A 144-nucleotide region that included residues of the PBS through the AUG hairpin was identified by this approach as the minimal sequence necessary and sufficient for genomic RNA dimerization during virus assembly169,170.
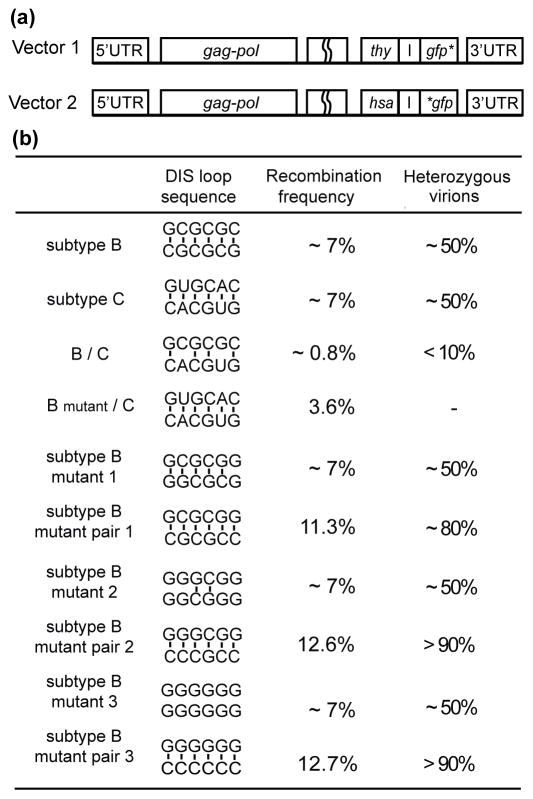
Early biochemical and mutagenesis studies led to proposals that dimerization is initiated by a conserved hairpin that serves as the dimer initiation site (DIS)48,51,74,171–175, and recent genetic and biochemical studies by Hu and co-workers have provided strong in vivo evidence in support of this mechanism176,177. They showed that, for viruses produced by cells dually infected with different HIV-1 subtypes (subtype B, DIS loop sequence GCGCGC; and subtype C, DIS loop sequence GUGCAC), the inter-subtype recombination frequency is much lower than intra-subtype recombination. The recombination rate increased four-fold by a 3 nucleotide substitution in the subtype B DIS, which made it fully complementary with the subtype C DIS176. Similar conclusions were reached in studies involving co-infection with mutants containing DIS sequences that were complementary to each other, but not self-complementary177, Figure 2. These results strongly argue that HIV-1 genomes dimerize prior to packaging, and that RNA-RNA recognition is mediated by the DIS.
Figure 2.
Results of recombination experiments used to assess RNA dimerization and heterozygote formation. (a) 293T cells were co-infected by two parental vectors containing 5′-UTR and gag-pol, followed by either thy or hsa reporter gene. Each vector was designed with a non-native gfp gene containing mutations either on the 5′- or the 3′-end. Therefore, only recombination between these two RNAs would generate functional gfp gene during reverse transcription in an infection event by these newly produced proviruses. (b) The recombination rate of HIV-1 subtype B sequences was approximately 7%, which was close to that of subtype C viruses. However, when cells were co-infected with both subtype B and C vectors, the inter-subtype recombination rate was only 0.8%. This may be explained by the fact that less heterozygous virus were formed. The inter-subtype recombination rate increased approximately 4-fold when the DIS of subtype B was substituted by subtype C DIS sequence176. Similar results were obtained in subtype B DIS mutants. When cells were co-infected with two plasmids containing the same non-self-complementary DIS sequence (mutant 1, mutant 2, or mutant 3), the recombination rates remained at the wild type level. Nonetheless, when cells were dual infected with DIS mutant pairs (mutant pair 1, mutant pair 2, or mutant pair 3), the recombination rates were significantly higher. It is conceivable that in these mutant pairs, formation of heterozygous virions is preferred, thus resulting in enhanced recombination frequencies177.
The incorporation of RNA into assembling particles was recently imaged in live cells by Bieniasz and colleagues15,16. In these studies, the genome RNA was engineered with multiple stem loops that bind to the bacteriophage MS2 coat protein, which could then be imaged in vivo upon binding to an MS2-NLS-GFP (GFP = green fluorescence protein) protein. The HIV-1 Gag protein was tagged with the mCherry fluorophore. The genome RNA and Gag proteins were then imaged in co-expressed living cells using dual-color total internal reflection (TIR) fluorescence microscopy. In the absence of Gag, genome RNA molecules were highly dynamic and did not localize at specific sites at or near the plasma membrane. However, in the presence of Gag, genomes were targeted to well-defined sites on the plasma membrane, initially exhibiting slow lateral movement and gradually becoming static. The targeting of the RNA molecules to plasma membrane assembly sites was achieved by a sub-detectable number of Gag proteins (likely ~12 or fewer). Detectable and increasing populations of Gag molecules appeared at the RNA localization sites after the RNA became stationary. The intensity of GFP-labeled RNA signal did not increase throughout this process, indicating that both RNA molecules are simultaneously trafficked to plasma membrane assembly sites.
The location in the cell where retroviral RNAs dimerize has been a subject of considerable recent interest. As indicated above, RSV genomes are likely to dimerize in the nucleus. Dimerization of the MoMuLV genome has also been proposed to occur within the nucleus, based in part on the observation of higher recombination frequencies observed for co-expressed MoMuLV-based vectors transcribed from physically proximal proviruses as compared with vectors expressed by spatially separated proviruses 178,179. In addition, co-expressed MoMuLV genome RNAs preferentially self-dimerize, resulting in a non-random co-packaging of homo- and heterodimeric genomes consistent with dimerization in the nucleus180,181. In contrast, HIV-1 co-packages co-expressed genomes as homodimers and heterodimers in random proportions, suggesting that dimerization may occur outside the nucleus 182,183. Very recently, Hu and co-workers developed a cell fusion system, in which two groups of cells (modified to help enable cell-cell fusion) were differentially infected with two strains of HIV-1, each containing mutations in Gag that cause a severe replication defect. When the two groups of cells are fused, the different Gag proteins can co-assemble, rescuing each other’s replication defect. Interestingly, viruses that assemble and bud from the fused cells contain heterozygous RNAs at levels similar to those obtained using a single cell co-infection system (based on a recombination assay), suggesting that HIV-1 RNA dimerization occurs in the cytoplasm and not in the nucleus184. In addition, cells were co-transfected with viral RNAs mutated to facilitate nuclear export by either the CRM-1 or nuclear export factor-1 (NXF1) dependent pathways. Different viral RNAs exported by the same pathway (CRM-1 or NXF1) were found to dimerize at levels similar to those observed for wild-type like RNAs, whereas differentially exported RNAs exhibited a reduced tendency to be packaged as heterodimers184. Thus, the findings obtained by different laboratories involving different retroviruses argue both for and against nuclear-associated genome dimerization. It is conceivable that dimerization occurs in multiple sites in infected cells, and the site that predominates depends on temporal or other conditional factors that have not been explored.
Structural Studies of the HIV-1 5′-UTR
Over the past 20 years, considerable effort has been made by many research groups to identify the RNA residues and structures that are responsible for genome selection and packaging. Most studies indicate that the primary packaging determinants reside within the 5′-UTR, and most structural studies therefore focused on this region of viral genomes. Deletion mutagenesis studies have shown that ~120 nucleotides located upstream of the Gag start codon are required for efficient packaging43–46,48–51. Although this region has sometimes been referred to as the Ψ-site, accumulating lines of evidence indicate that efficient HIV-1 genome packaging requires nearly the entire 5′-UTR and possibly extends into the gag coding region9,54,59,185. Thus, unlike for some other retroviruses, a minimal packaging element that is independently capable of directing efficient HIV-1 genome packaging has yet to be identified and independently validated.
Electron microscopy (EM) studies of RSV, MoMuLV and some other tumor viral RNAs purified from sucrose gradient sedimentation revealed that residues near the 5′-end of the RNAs (ca. 300–500 nucleotides) adopt X- or Y- shaped dimer linkage structure (DLS)29,32,65,186,187. Circular loop structures without free 5′-ends were observed in the EM studies of the HIV-1 genome RNAs extracted from virions188. It is worthy to note that the RNA samples used in this study were treated by 50% formamide with 2.5 M of urea in order to unwind the tightly packed coil structures, suggesting that the dimer linkages are highly stable. Similar loop structures were observed in a NC-incubated RNA (1–744nt) prepared by in vitro transcription71. These findings were interpreted in terms of two dimer interfaces near the very 5′-end of the RNA.
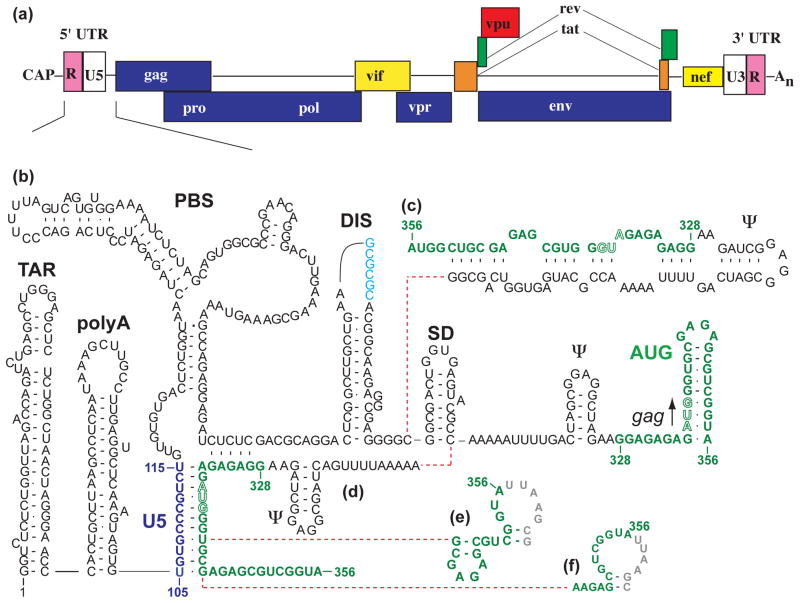
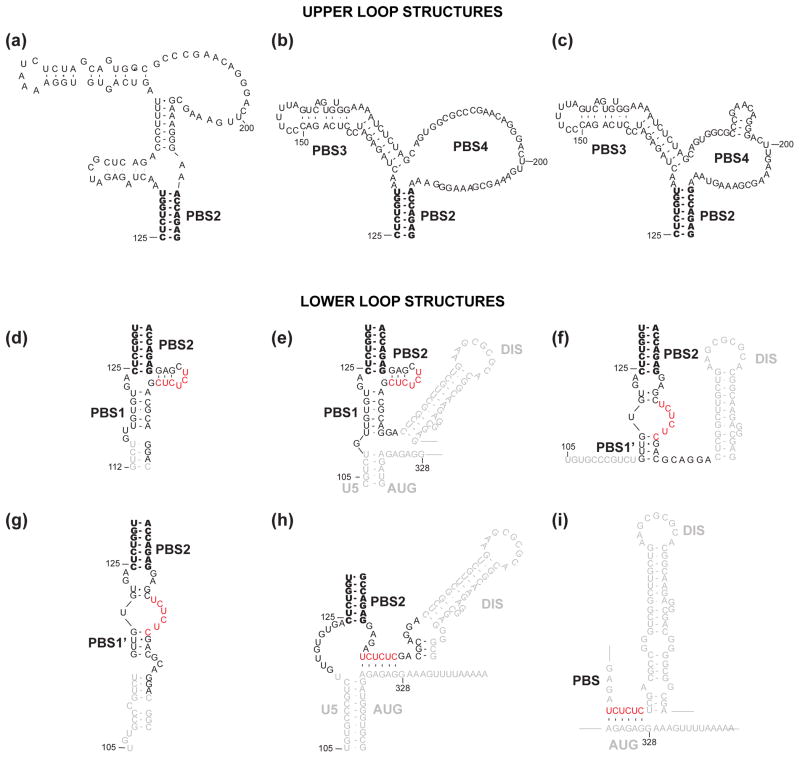
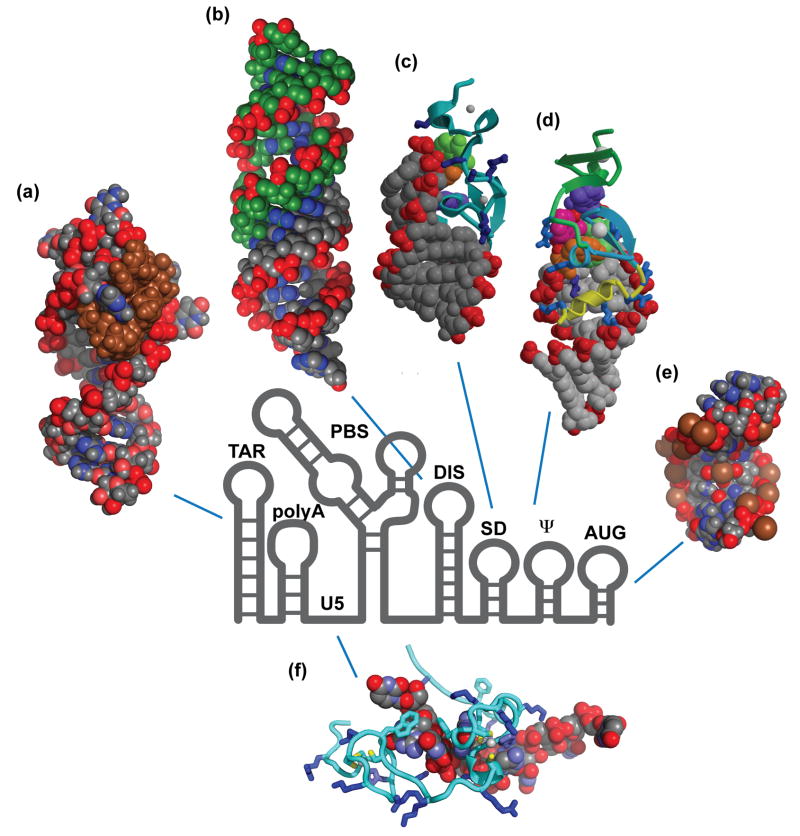
More than 20 different secondary structures have been predicted for the HIV-1 5′-UTR over the past 25 years47–50,69–71,73–75,189,190. The collection of site-directed mutagenesis experiments, chemical and enzymatic accessibility assays, phylogenetic studies, and free energy calculations are generally consistent with a 5′-UTR structure that consists of a series of stem-loop structures connected by relatively short linkers, Figure 3. These stem-loops are (from 5′ to 3′): trans-activation region (TAR), the 5′ polyadenylation signal (polyA), the primer binding site (PBS), the dimer initiation site (DIS), the major splice donor (SD), and the Ψ hairpin. The chemical probing data for these regions are not entirely self-consistent, and for other regions of the 5′-UTR, significant variations in nucleotide accessibility have been reported. Results of nucleotide accessibility mapping experiments highlighting similarities and differences among the chemical probing results obtained for these regions of the 5′-UTR are summarized in Figure 5. Based in part on these variations, five different models have been proposed for the AUG region over the past eight years alone, Figure 3b-f, and six different models have been proposed for the PBS loop, Figure 4. In the following subsections, experimental data and modeling results are summarized according to the current understanding of structure/function relationships.
Figure 3.
(a) Diagram of the composition of the HIV-1 genome showing locations of the 5′-UTR and splice sites. (b)-(f) Representative secondary structures predicted for the HIV-1 5′-UTR. In this figure, variations among the recent predictions for the AUG region (green) are shown.
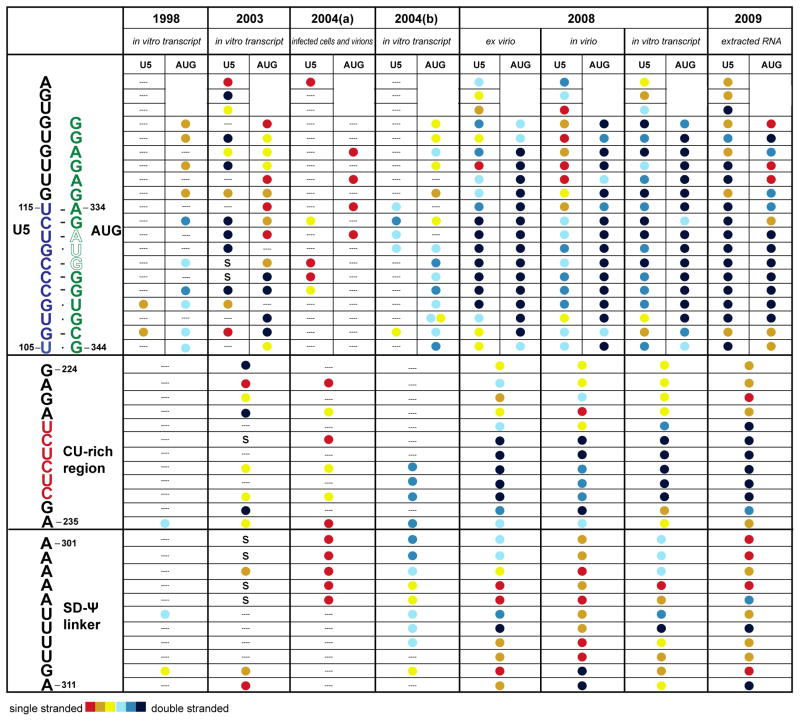
Figure 5.
Summary highlighting the variations in chemical/enzymatic probing results obtained for nucleotides in U5:AUG, CU rich region (CU RR) and the linker between the SD and Ψ hairpins in the context of the full length HIV-1 5′-UTR (Red and dark blue circles denote residues predicted to exist in single-stranded and double-stranded secondary structures, respectively; “S” denotes transcription termination points; dashes denote no data reported). 1998: Damgaard et al.232 enzymatically modified the in vitro transcribed HIV-1 1–401 RNA. 0.1–0.2 pmol/μl RNA was first renatured by incubating it at 80°C for 5 min in 10 mM HEPES-KOH (pH 7.6), 100 mM KCl, followed by slow cooling. The RNA was then digested by RNase T1, RNase T2 and RNase V1 for 20 min on ice. 2003: Abbink et al.234 analyzed the in vitro transcript of the complete HIV-1 5′-UTR plus some of the Gag ORF (−454-+376) by DMS and kethoxal. 10 μg RNA was treated at room temperature with kethoxal for 10 min and DMS for 5 min with a concentration of 100 mM sodium cacodylate and 1 mM MgCl2. 2004(a): Paillart et al.62 directly analyzed the HIV-1 HXB2 RNA conformation in infected cells and in viral particles using DMS. Cells and viruses were treated with 3 μL DMS at 37°C for 4 and 8 min. 2004(b): Damgaard et al.71 probed the in vitro transcribed 5′ 744 nucleotides conformation of the HIV-1 HXB2 genome. ~20 μg/μl RNA was digested by RNase T1, T2 and V1 respectively in 100 mM KCl, 2 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 10% glycerol and 0.18 Unit/μl RNasin. 2008: Weeks and coworkers104 applied hSHAPE to study the first 900 nucleotides of the HIV-1 pNL4-3 RNA genome under biological relevant conditions (ex virio, in virio and in vitro transcripts). The RNA is treated with NMIA under 37C for 50 min. The color scheme was approximated based on the raw data listed in the supplementary material104. 2010: Weeks and co-workers262 used the same technique to study the whole HIV-1 genome secondary structure. ~0.11 uM HIV-1 genome was modified in 50 mM HEPES, 20 mM potassium acetate and 3mM MgCl2 by NMIA at 37°C for 15 min. (DMS modifies adenines at position N-1 and cytosines at N-3 when A and C are not base-paired; RNase T1 preferentially cleaves at the 3′ side of the unpaired Guanine bases; RNase T2: predominantly cleaves single-stranded RNA; RNase V1 preferentially cleaves double-stranded or stacked RNA; Kethoxal specifically cleaves single-stranded guanidines; NMIA modifies the ribose backbone of the unpaired bases; hSHAPE = selective 2′-hydroxyl acylation analyzed by primer extension).
Figure 4.
Secondary structures predicted for the upper (a–c) and lower (d–i) loops of the HIV-1 PBS. Nearly all the models include a common base paired helix in the center of the PBS (PBS2, in bold). Upper loop predictions: (a) Berkhout and co-workers predicted a structure containing an apical hairpin ending in an A-rich loop199 for the HIV-1 HXB-2 RNA genome. (b) Damgaard et al. proposed that the important A-rich sequence is displayed in an internal loop232. (c) Wilkinson and co-workers further predicted a small hairpin structure in the PBS4 region, based on the SHAPE probing104 for the HIV-1 NL4-3 RNA genome. Lower loop predictions: (d,e) Kjems and co-workers predicted that the CU rich region (red) forms a small hairpin with the nucleotides right after the PBS2 stem loop71,232. (f,g) Berkhout and co-workers proposed that CU rich region forms a loop structure199,227. (h) Weeks and co-workers predicted that the CU rich region forms a long-range interaction with the AG rich linker between Ψ and AUG104,262, consistent with an earlier prediction by Lever and co-workers72 (i).
TAR
There has been considerable interest in the TAR domain of the 5′UTR due to its essential role in Tat-mediated transcriptional activation191–194, but other roles in replication have also been proposed, including roles in dimerization71, strand transfer during reverse transcription171, as a possible HIV-1 derived miRNA during latency184, and packaging54,55,195. Mutagenesis studies have sometimes been difficult to interpret due to the dominant negative effects that the mutations have had on transcription. For example, Berkhout and co-workers showed that mutations designed to disrupt the structure of TAR lead to significant reductions in genome packaging efficiency195 and concluded that the TAR hairpin structure shown in Figure 3b is important for genome packaging. Similar results have been reported by Clever and co-workers196. However, efficient genome packaging and replication was subsequently demonstrated for an HIV-1 variant, in which the TAR hairpin and Tat gene is substituted by a tetracycline-inducible tetO-rtTA system197, indicating that the TAR hairpin is required for efficient transcription and replication but does not play an essential role in packaging198.
Chemical probing, mutagenesis, and other structural studies are in general agreement that the 57-nucleotide TAR sequence forms the stable hairpin shown in Figure 3b71,104,199. The TAR hairpin contains a conserved 3-nucleotide pyrimidine bulge that binds to the viral Tat transactivator protein200–203 and an apical 6-nucleotide loop that binds to the cyclin T1 subunit of the cellular transcriptional elongation factor (pTEFb)204–206. The high-resolution 3D solution NMR structure of the upper portion of the TAR stem loop has been determined in both the absence and presence of the bound ligands155,200,207–218, Figure 6a. The unbound Tat binding 5′-bulge residues are conformationally flexible, but become rigid upon binding to Tat-derived peptides. Binding is mediated by an arginine side chain (Arginine fork)216,219, which stabilizes a coaxially aligned TAR conformation by interacting with residues at the interface that connects the lower and upper A-form helical segments200,207,208,217,218. The 5′-uracil of the bulge forms a base triple with the adjacent A-U base pair, causing a significant distortion of the RNA backbone. The intrinsic conformational mobility of the bulge plays an important role in ligand binding220–224, and information about local dynamics trajectories and transiently formed structures has facilitated the identification of new inhibitors that bind to the Tat binding pocket and inhibit Tat-mediated activation and viral replication221.
Figure 6.
Representative high-resolution 3D structures determined by NMR or X-ray crystallography for relatively small RNA fragments of the HIV-1 5′-UTR (P,O atoms colored red; C, N atoms gray and blue, respectively, except as noted). Shown are (a) the NMR structure of the TAR hairpin bound to a cyclic peptide mimic of the Tat protein (brown)276, (b) crystal structure of the DIS:DIS kissing complex (C atoms of the two RNA molecules shown in green and gray)243, (c) the NMR structure of the SD hairpin bound to NC (backbone displayed as a ribbon; basic side chains displayed in blue color)253, (d) the NMR structure of the Ψ hairpin bound to NC (in ribbon format)132, (e) the NMR structure of the AUG hairpin (refined with a force field that included sodium ions, colored brown)260, (f) the NMR structure of the NC protein (ribbon format, with basic residues shown in blue) bound to a U5 oligoribonucleotide137.
PolyA
Residues immediately following the Tat hairpin have been predicted to form a second hairpin, called polyA, which contains the AAUAAA polyadenylation signal in an unstructured loop190. This sequence is part of the repeat (R) element that is duplicated near the 3′-end of the viral transcript. Although the 3′ polyadenylation signal is known to function in mRNA maturation, little is known about the function of the polyA hairpin located in the 5′-UTR. Parslow and coworkers found that mutations disrupting the poly(A) base pairing caused profound defects in both genome packaging and viral replication196. Compensatory mutations that restored base pairing also restored genome packaging, leading to conclusions that the secondary structure of PolyA hairpin, and not its primary sequence, is important for packaging196. Berkhout and coworkers similarly found that destabilization of the polyA hairpin results in diminished genome packaging225. However, the Berkhout lab later showed that destabilizing the poly(A) hairpin can lead to significant reductions in the intracellular levels of HIV RNA, and this can account for the observed reductions in packaging. Their results further suggested that the polyA hairpin is required for both efficient repression of the 5′-polyadenylation site and full activation of the 3′-polyadenylation signal, but may not be essential for packaging (although a 5′-terminal hairpin seems to be necessary)226.
To date, no 3D structural information is available for residues of the polyadenylation signal. In addition to the proposed hairpin structure shown in Figure 3b, residues in this region have been proposed to form base pairs with DIS in the long-distance interactive (LDI) model of Berkhout and co-workers227. Paillart and co-workers suggested on the basis of a phylogenetic analysis and biochemical studies that the loop residues of the polyA hairpin base pair with residues in the matrix coding region of the genome, forming a long range pseudoknot228. This proposed interaction is conserved in all HIV-1 isolates as well as in HIV-2 and simian immunodeficiency virus228.
PBS
Residues of the PBS region of the 5′-UTR play a critical role in replication by serving as the binding site for the human tRNALys RNA, the primer for reverse transcription, and serving as regulatory elements that affect the initiation of reverse transcription208,229–231. Numerous secondary structural models have been proposed for residues of the PBS, based on chemical and enzymatic probing, computer modeling, and mutagenesis experiments71–73,104,199,205,213,227,231–234, Figure 4. In the upper loop, some of the structures contain an apical hairpin ending in an A-rich loop, which is not supported by the phylogenetic analysis. Kjems and co-workers instead proposed that the important A-rich sequence is displayed in an internal loop which is conserved in all major subtype isolates of HIV-171, Figure 4a. The bottom region of PBS has more variations with the most dramatic difference in the CU rich region, Figure 4b. In spite of the significant discrepancy in the top and bottom part of PBS, nearly all models include a common base paired helix PBS2 in the center of the PBS, Figure 4.
The Lever group first predicted that the CU rich region forms a long-range interaction with the AG rich linker between the Ψ and AUG hairpins72, and supporting evidence has more recently been obtained by mutagenesis235 and SHAPE probing104,233, Figure 5. Kjems and coworkers predicted that the CU rich region forms a small hairpin with the nucleotides right after the PBS2 stem loop9. However, the Berkhout group interpreted chemical reactivity of the CU rich region in terms of a loop conformation, Figure 5. To identify the contribution of the PBS region to genome packaging, Parslow and co-workers57 created a series of 13 disruptive and compensatory mutations within this region using a single-round replication assay. In agreement with the results from the Berkhout group213 using spreading-virus assay, the top part of PBS had little functional effect on genome packaging. However, although mutations designed to disrupt the predicted base pairing in the lower stem impaired replication, compensatory mutations intended to restore its structure exacerbated the defect. Mutations targeting the middle stem PBS2 by both groups confirmed its biological role in genome packaging. However, compensatory PBS2 mutations designed by Berkhout group failed to restore replication while Parslow group’s mutations corrected the packaging defect and partially restored the viral infectivity. This discrepancy may have resulted from different mutation sequences or using different assays. Although there is currently little agreement regarding the structure of the PBS, all studies consistently indicate that some of the PBS residues play an important role in genome packaging.
DIS
The DIS has also received considerable attention, with nearly all studies supporting the hairpin structure shown in Figure 3b. The DIS promotes dimerization via formation of intermolecular “kissing contacts” involving the GC-rich palindromic loops74,236, Figure 3b. As indicated above, co-infection with plasmids that encode non-self-complementary DIS loops that are complementary to each other has been used to enhance packaging of heterodimeric RNAs177. Although DIS-containing oligo-RNAs are capable of converting from a kissing dimer to an extended duplex, larger RNAs that include the entire 5′-UTR and the first 265 nucleotides of the gag coding region do not appear to form an extended duplex, and it is likely that the kissing dimer is the biologically relevant species173. Mutations designed to disrupt the DIS structure, or deletion of the entire DIS stem-loop, inhibit RNA dimerization in vitro and lead to significant reductions in both genome packaging and infectivity in vivo48,51,53,74,171–175,193,210. Multiple secondary structures have been predicted for residues below the GG bulge, Figure 3b71,104. Although DIS is important for genome packaging, it does not appear to bind with high affinity to the NC protein237, and it is conceivable that, as observed for the MLV packaging signal66, DIS-dependent HIV-1 genome dimerization promotes exposure of high-affinity NC binding sites.
The structure of the kissing dimer has been determined by NMR74,237–242 and crystallography243,244, Figure 6b. Some discrepancies are apparent upon comparison of the structures. The NMR solution structure shows a symmetrical homodimer where the averaged loop plane is approximately perpendicular to the axis of the stems, whereas a perfect coaxial alignment with the stems was observed in the crystallography structure. Unlike the NMR structure where all bases are inside the RNA molecule, the crystallographic structure presents the two loop adenines as stacked in a bulged out conformation. A recent NMR structure of the kissing dimer consisted of two coaxially aligned hairpins with loop adenines in a bulged in conformation239. The kissing dimer can be converted into a more thermodynamically stable extended dimer by incubation at 55°C245–247, or by incubation at lower temperatures in the presence of NC106,248,249, and X-ray and NMR structures of the extended dimer have also been solved246,247. However, as indicated above the biological relevance of this species remains uncertain. Marino and co-workers demonstrated a proton-coupled dynamic conformational switch in the kissing complex. The NC catalyzed maturation of the kissing complex was shown to directly correlate to the observed proton coupled dynamics242. The structure237,250 and dynamics251 of the lower stem and bulge have also been examined.
We note here that a second dimer interface that is separate and distinct from the DIS was proposed on the basis of EM studies (see above). Based on computer-modeling analysis, polyA was proposed to be the second DLS188. However, deletions and mutations of the proposed dimer-promoting, palindromic AAGCUU element in PolyA were found to have no impact on genome dimerization58,252. More recent biochemical studies with oligo-RNAs indicate that TAR has a high propensity to dimerize and is capable of promoting dimerization in RNAs that lack the DIS sequence, and it was therefore suggested that TAR may contribute to a second DLS71. However, as indicated above, TAR can also be deleted or substituted by a non-viral transcriptional activator without significantly affecting RNA packaging or infectivity. Thus, the second dimerization site observed in the EM images either has yet to be properly identified, or its role in genome packaging not significant.
SD
Results of nucleotide accessibility mapping are generally consistent with the SD hairpin structure shown in Figure 3b. The major splice donor site is located in the GGUG loop of the proposed hairpin. The isolated SD hairpin is capable of binding NC with high affinity253. The three-dimensional structure has been determined for the isolated DIS hairpin253 and its complex with NC133 have been determined by NMR methods, Figure 6c. High affinity binding is mediated by interactions between the zinc knuckles of NC and exposed guanosines in the GGUG tetraloop (as also observed in a NC:Ψ hairpin structure; see below)133. Interestingly, mutations designed to disrupt the base pairing in the lower stem of the hairpin did not affect genome encapsidation or replication, leading to suggestions that this hairpin does not play a role in genome packaging48,49.
Ψ
Most evidence indicates that the hairpin structure predicted for the Ψ residues (Figure 3b) plays an important role in genome packaging – hence its name. Hayashi et al reported that a fragment of the 5′-UTR containing the Ψ hairpin is capable of directing the packaging of heterologous RNAs into virus-like particles69, but this finding has not been independently verified. Russell and co-workers reported that the Ψ hairpin and downstream GA rich region are required not only for packaging but also for genome dimerization235. Deletion of Ψ, and mutations designed to disrupt base pairing in the stem, have been reported to lead to significant reductions in genome packaging47,49,59,235, whereas compensatory mutations designed to restore base pairing in the stem47, and the complete substitution of Ψ by a different NC-binding RNA fragment254, largely restored packaging efficiency.
RNAs containing Ψ bind NC with high affinity75,133,211, and high-resolution NMR structures have been determined for the free Ψ hairpin255 and for a high affinity NC:Ψ RNA complex132, Figure 6d) Both of the HIV-1 NC zinc knuckles contain a hydrophobic pocket that interacts specifically with the bases of exposed guanosines130,132,133,137. For both zinc knuckles, the guanosine nucleobase inserts deeply into the hydrophobic pocket and forms hydrogen bonds with backbone NH and carbonyl groups of the polypeptide. The ability to bind specifically and tightly to exposed guanosine bases was proposed to be a primary function of the HIV-1 NC zinc knuckles132,133. Surprisingly, although mutations that disrupt the stem structure of the Ψ hairpin severely impair genome packaging, substitution of the NC binding GGAG loop by GCUA or AAGA did not significantly affect packaging or replication235. Both substitutions contain a tetraloop guanosine, and oligoribonucleotide Ψ RNAs containing these mutations are capable of binding NC (albeit with weaker affinities of ~ 4 μM and ~ 800 nM, respectively) (Heng and Summers, unpublished results). The single zinc knuckle domain of the MoMuLV NC protein was also found to bind exposed guanosines66,136, but only one of the two RSV NC zinc fingers appears to function as a guanosine binding site upon binding to a minimal RNA packaging element135. Thus, guanosine recognition does not appear to be a universal function of the highly conserved structural motif.
AUG
Residues that span the gag start codon (here called AUG) have been proposed to adopt a number of different secondary structures, Figure 3b49,256,257. Early nucleotide probing, mutagenesis studies and free energy predictions were generally consistent with the hairpin structure shown in Figure 3b48,59,73. Studies of AUG-derived oligoribonucleotides indicated that the hairpin contains an unstable stem and a stable GNRA tetraloop (N: any base; R: G or A)258–260, and the 3D structure of the hairpin RNA was solved by NMR methods260, Figure 6e. GNRA tetraloops have often been found in nature to participate in long range RNA-RNA interaction214,261, and it was therefore hypothesized that the GNRA loop of AUG might interact with the other parts of the HIV-1 5′-UTR261. More recently, using a combination of phylogenetic analyses, biochemical probing, mutagenesis, and computer-assisted RNA modeling, Berkhout and co-workers suggested that AUG might instead base-pair with residues of the Unique-5′ element (U5) that connects the poly(A) and PBS stem loops234, Figure 3c. Supporting evidence for U5-AUG base pairing was obtained by Kjems and co-workers in enzymatic probing studies of a relatively large 5′-fragment of the HIV-1 genome (residues 1–744), combined with a newer computational RNA structure prediction algorithm71. However, subsequent nucleotide reactivity probing of viral RNAs in transfected cells and intact virions by Ehresmann and co-workers revealed that several nucleic acid bases predicted to participate in U5:AUG interactions were readily modified by chemical probes, indicating that they are exposed and highly reactive, a finding that is not compatible with the proposed U5:AUG base pairing62. More recently, results of SHAPE experiments employed by Weeks and co-workers were used to argue that residues of AUG do, in fact, base pair with residues of U5, forming a stable structure that persists in transfected cells, in virions, and in in vitro transcribed 5′-UTR RNAs104 (although the proposed base pairing pattern differed somewhat from the predictions by the Kjems and Berkhout laboratories). The SHAPE method is purportedly more sensitive to unstructured elements than traditional chemical probing techniques, and it is therefore unclear why the residues of U5 and AUG were reactive to traditional chemical probes but poorly reactive to the SHAPE probes. All of the in vivo chemical probing experiments were conducted using transfected cells and virus particles under what appear to be similar, native-like conditions. The AUG structure was revised in a subsequent paper by the Weeks group262, although the SHAPE reactivities of the participating residues did not appear to change relative to the earlier studies.
Larger Fragments of the 5′-UTR
Although early studies showed that a relatively small region of the HIV-1 5′-UTR containing the Ψ hairpin is sufficient to direct the packaging of heterologous RNAs into virus-like particles69, more recent studies showed that mutations in the stem of Ψ designed to disrupt base pairing, or substitution of the NC-binding GGAG tetraloop by AAGA, resulted in only modest reductions in packaging47, and it is now clear that a much larger portion of the 5′-UTR participates in genome recognition and packaging. Unfortunately, 3D structural information for larger, multi-hairpin RNAs remains sparse. High-resolution structural studies of RNA by X-ray crystallography can be problematic, due in part, to conformational heterogeneity and a relatively uniform, negative surface charge that can hamper crystallization. NMR has its own issues: NMR signal degeneracy, spin relaxation, and a general paucity of long-range restraint information have limited the utility of NMR for studying even modest-sized RNAs. As of 2010, the RNA structures deposited in the Nucleic Acid Structure Database comprise only 25 nucleotides on average, and only three structures comprise more than 50 nucleotides263. For these reasons, structural studies of larger RNAs, including the intact 5′-UTR, have relied primarily on nucleotide protection, mutagenesis, and biochemical experiments62,71,73,79,104,199,233,234.
RNA cross-linking has been utilized to probe for long-range RNA-RNA interactions, and Kjems and co-workers used a UV cross-linking approach to identify a novel tertiary interaction within the PBS hairpin structure71. More recently, Fabris and co-workers used mass spectrometry and chemical cross linking to probe for potential long-range interactions in a 5′-UTR fragment encompassing residues of the DIS through the AUG hairpins264, and a 3D model was generated using the results of chemical crosslinking as distance restraints. These studies indicated that the 5′-UTR fragment forms a globular structure, in which the GAGA tetraloop of AUG participates in A-minor like interactions with the stem of DIS (a common interaction for this class of tetraloops258,261). Of course, in the context of the intact 5′-UTR, it is certainly possible that AUG could instead base pair with U5 or participate in other predicted long-distance interactions (see below). No other high-resolution 3D structural information for larger fragments of the 5′-UTR has been published to date.
The 5′-UTR Can Adopt Multiple Conformations – A Regulatory Role?
There is considerable evidence that the HIV-1 genome, and its 5′-UTR, can adopt multiple conformations. Even the earliest nucleotide probing experiments by the Ehresmann group suggested that the dimeric form of the 5′-end of the viral genome (residues 1–500) likely adopt multiple distinct and mutually exclusive structures that could modulate the function of the 5′-UTR73. The isolated, recombinant HIV-1 5′-UTR (residues 1–373) appears to form a mixture of monomers and dimers, and native gel electrophoresis studies revealed intermediate bands after incubated for 8hr at 55°C in dimer formation buffer (50 mM Tris-HCl, pH 7.5, 10mM KCl, 1mM MgCl2)265. Berkhout and co-workers showed that a 1–290 nucleotide fragment of the HIV-1 5′-UTR can adopt two distinct monomeric conformations that migrate at different rates on native polyacrylamide gels266. The fast migrating species was proposed to form a rod-like structure (termed LDI, for “Long Distance Interactions”)267 based on site-directed mutagenesis and secondary structure calculations267, Figure 3f. Addition of Mg2+ shifts the equilibrium towards the slowly migrating species, which is referred to as the “branched multiple hairpin” (BMH) conformer266. A longer fragment (1–744) of the HIV-1 genome is also capable of forming LDI and BMH conformations71. The NC protein can efficiently promote dimerization of the BMH conformer, but not the LDI species, and the burial of the DIS loop in the LDI was proposed to inhibit dimerization267. The equilibrium of these two species was hypothesized to regulate dimerization, translation and genome packaging268,269. However, mutations designed to shift the LDI-BMH equilibrium did not significantly affect translation efficiency227. No evidence for an LDI-like species was observed by in vivo nucleotide probing, although the presence of a minor LDI species would likely be difficult to detect by this approach62.
There is also evidence suggesting that the structure of the genome changes as the virus matures. Genomes isolated from mature HIV-1 viruses under mildly denaturing conditions exist as a mixture of monomeric and dimeric species (80–95% dimer)168, and the proportion of dimers can be reduced to ~ 40–50% by inactivating the protease105, the distal NC zinc fingers252 or the DIS52,210. The electrophoretic mobility of the dimeric genome increases as the virus ages (from 2hr to 9hr). In addition, dimers in newly released viruses are thermolabile, and can dissociate during extraction270, but after ~ 4-6hr the dimers become “mature” and are thermostable168. Thus, both the monomer/dimer ratio and the stability of the dimer appear to vary as a function of the age of viruses168. In this regard, the conclusions based on SHAPE experiments that the 5′-UTR adopts a single structure in cells, in virions, and in vitro104, seems surprising.
Overview and Future Directions
A number of findings made over the past five years or so have significantly advanced our understanding of the mechanism of HIV-1 genome packaging. Fluorescence imaging studies have enabled real-time visualization in living cells of RNA trafficking, the assembly of Gag proteins, and the recruitment of cellular proteins during virus assembly15,16,271–273. These studies have shown that genome trafficking to virus assembly sites (from unknown origins in the cell) is dependent on Gag, that virions do not assemble repeatedly at pre-formed assembly sites, that two copies of the genome are simultaneously trafficked to the membrane, that the RNA-Gag complex contains a small number of Gag proteins (probably ~12 or fewer), and that additional Gag molecules are recruited for assembly subsequent to the docking of the initial Gag:RNA complex on the plasma membrane. Although small amounts of monomeric genomes can be isolated from young HIV-1 particles under mildly denaturing conditions, the findings from fluorescence imaging experiments, together with recent results of genetic recombination experiments, provide convincing evidence that HIV-1 genomes are selected for packaging as dimers.
A significant outstanding questions is: Where in the cell does Gag:RNA recognition occur? Over the past five years or so, Parent and colleagues have provided compelling evidence that genome packaging by RSV Gag is dependent on the shuttling of Gag through the nucleus, and that this process is mediated by the NLS and NES activities of Gag’s MA domain. Although Green and co-workers presented early evidence that HIV-1 Gag transiently accesses the nucleus prior to assembly, and that mutations in the MA domain of Gag that interfere with this process lead to aberrant genome packaging, the role of HIV-1 Gag in nuclear export of the viral genome remains both controversial and un-validated. By engineering the RNA to utilize different nuclear export pathways, Hu and co-workers have obtained evidence that dimerization of the HIV-1 genome probably takes place in the cytoplasm (although dimerization does appear to depend, at least to some extent, on the mechanism of nuclear export)184. Assuming that HIV-1 Gag binds preferentially to the dimeric genome (as observed for MoMuLV, but not yet reported for HIV-1), this finding would seemingly favor a packaging mechanism in which HIV-1 Gag:RNA assembly is initiated in the cytoplasm.
Regardless of the location of Gag:RNA recognition, there remains considerable interest in understanding the molecular basis for genome selection. How does Gag specifically direct the packaging of the dimeric, unspliced genome? Although the MA domain of Gag could play a direct role by binding tightly and specifically to the genome, such a mechanism has yet to be firmly established by experiment. Since MA plays known roles in intracellular trafficking, an alternative role would be to help direct Gag to sites in the cell where the viral genomes are located. On the other hand, there is considerable experimental evidence that the NC domain of Gag plays a direct role in genome selection by binding tightly to recognition elements in the viral RNA. In the case of MoMuLV, it appears that dimerization of the genome, which may be induced by the chaperone activity of NC, exposes about a dozen high affinity NC binding sites that were sequestered and unable to bind NC in the monomer, and it was suggested that dimerization of the MoMuLV 5′-UTR leads to formation of an RNA structure that exposes unstructured UCUG NC binding elements66,67,274. Similar conclusions were reached recently by SHAPE-based in virio chemical probing and mutagenesis experiments 68. The RNA binding properties of a larger MoMuLV Gag fragment differ from those of the isolated NC domain68, and quantitative NC binding and 3D structural studies of such complexes are therefore also warranted.
High-resolution NMR structures have been reported for several isolated RNA hairpins corresponding to functionally important elements within the HIV-1 5′-UTR, and for the most part, the structures are consistent with secondary structure predictions made on the basis of chemical accessibility and phylogenetic mapping, mutagenesis and biochemical studies, and free energy calculations. NC:RNA structures have been determined for two of the predicted hairpins (SD and Ψ) and for a single-stranded element within the U5 region of the 5′-UTR, revealing details about atomic-level interactions that are important for high-affinity NC:RNA binding. However, none of these RNA elements appear to be strictly required for packaging, and because these studies involved relatively small fragments of the 5′-UTR, they did not lead to insights into the mechanism of dimeric genome selection. To date, quantitative thermodynamic studies of the dimerization-dependent NC binding behavior of the HIV-1 5′-UTR (or other larger portions of the genome) have not been reported, and in this regard, it should be of interest to determine if the HIV-1 5′-UTR exhibits dimerization-dependent NC (or Gag) binding behavior similar to that observed for the MoMuLV 5′-UTR.
There is also a need to develop better approaches for structural studies of large RNAs that are conformationally heterogeneous, as appears to be the case for the HIV-1 5′-UTR. The use of chemical probing and nucleotide cross-linking experiments has provided insightful structural information for some regions of the 5′-UTR, but for other regions, inconsistencies among the experimental data have led to a variety of structural predictions and mechanistic interpretations. These inconsistencies could potentially be due to the presence of multiple RNA structures and conformational heterogeneity, which could complicate interpretation of bulk reactivity data. For example, a 100 nucleotide RNA corresponding to the MoMuLV core encapsidation signal was shown by NMR to exist under physiological salt conditions as a mixture of four minor monomeric conformers, two minor dimeric conformers, and several minor higher-order multimers, in addition to the major dimeric species274, and this was offered as a potential explanation for the significant differences observed between a dimer model derived by chemical probing275 and the NMR-derived structure274. Conformational heterogeneity is likely to present problems for any RNA structure determination method that cannot distinguish between multiple equilibrium species. New methods that combine NMR-derived high-resolution local structural information, which can distinguish between slowly interconverting RNA conformers, with low-resolution global structural information derived by single molecule cryo-Electron Tomography274 should prove useful for future studies of larger and functionally relevant fragments of the HIV-1 genome.
Acknowledgments
Support from the NIH (R01 GM42561, AI30917) is gratefully acknowledged.
Abbreviations used
- AIDS
acquired immunodeficiency syndrome
- BMH
branched multiple hairpin
- CRM-1
chromosome region maintenance-1
- LDI
long distance interaction
- HIV-1
human immunodeficiency virus type-1
- MoMuLV
Moloney murine leukemia virus
- NC
nucleocapsid protein
- NMR
nuclear magnetic resonance
- NXF1
nuclear export factor-1
- UTR
untranslated region
- ITC
isothermal titration calorimetry
- PBS
primer binding site
- PM
Plasma Membrane
- RT
reverse transcriptase
- SHAPE
selective 2′-hydroxyl acylation analyzed by primer extension
- VLP
virus-like particle
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rein A. Retroviral RNA packaging: A review. Arch Virology. 1994;9:513–522. doi: 10.1007/978-3-7091-9326-6_49. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr Top Microbiol Immun. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 3.Greatorex J, Lever A. Retroviral RNA dimer linkage. J Gen Virol. 1998;79:2877–2882. doi: 10.1099/0022-1317-79-12-2877. [DOI] [PubMed] [Google Scholar]
- 4.Paillart JC, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of retroviral genomic RNAs: Structural and functional implications. Biochimie. 1996;78:639–653. doi: 10.1016/s0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 5.Jewell NA, Mansky LM. In the beginning: genome recognition, RNA encapsidation and the initiation of complex retrovirus assembly. J Gen Virol. 2000;81:1889–1899. doi: 10.1099/0022-1317-81-8-1889. [DOI] [PubMed] [Google Scholar]
- 6.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: An inseparable pair. Nature Reviews Microbiology. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 7.Russell RS, Liang C, Wainberg MA. Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology. 2004 September;1(2) doi: 10.1186/1742-4690-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greatorex J. The retroviral RNA dimer linkage: different structures may reflect different roles. Retrovirology. 2004 August;1(18) doi: 10.1186/1742-4690-1-22. edition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Souza V, Summers MF. How retroviruses select their genomes. Nature Reviews Microbiology. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 10.Poon DTK, Wu J, Aldovini A. Charged amino acid residues of human immunodeficiency virus type-1 nucleocapsid P7 protein involved in RNA pacakaging and infectivity. J Virol. 1996;70:6607–6617. doi: 10.1128/jvi.70.10.6607-6616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cimarelli A, Sandin S, Hoglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J of Virology. 2000;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S-W, Aldovini A. RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes. J Virol. 2002;76:11853–11865. doi: 10.1128/JVI.76.23.11853-11865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muriaux D, Mirro J, Harvin D, Rein A. RNA is a structural element in retrovirus particles. Proc Natl Acad Sci USA. 2001;98:5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muriaux D, Mirro J, Nagashima K, Harvin D, Rein A. Murine leukemia virus nucleocapsid mutant particles lacking viral RNA encapsidate ribosomes. J Virol. 2002;76:11405–11413. doi: 10.1128/JVI.76.22.11405-11413.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci U S A. 2009;106:19114–9. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454:236–240. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogt VM. Retroviral virions and genomes. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Vol. 1. Cold Spring Harbor Laboratory Press; Plainview, N.Y: 1997. pp. 27–69. [PubMed] [Google Scholar]
- 18.Hu W-S, Temin HM. Genetic consequences of packaging two RNA genomes in one retroviral particle: Pseudodiploidy and high rate of genetic recombination. Proceedings of the National Academy of Sciences USA. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu WS, Temin HM. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 20.DeStefano JJ. Kinetic analysis of the catalysis of strand transfer from internal regions of heteropolymeric RNA templates by human immunodeficiency virus reverse transcriptase. J Mol Biol. 1994;243:558–67. doi: 10.1016/0022-2836(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 21.King SR, Duggal NK, Ndongmo CB, Pacut C, Telesnitsky A. Pseudodiploid genome organization aids full-length human immunodeficiency virus type 1 DNA synthesis. J Virol. 2008;82:2376–84. doi: 10.1128/JVI.02100-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coffin JM. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979;42:1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- 23.Onafuwa-Nuga A, Telesnitsky A. The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol Mol Biol Rev. 2009;73:451–80. doi: 10.1128/MMBR.00012-09. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gratton S, Cheynier R, Dumaurier MJ, Oksenhendler E, Wain-Hobson S. Highly restricted spread of HIV-1 and multiply infected cells within splenic germinal centers. Proc Natl Acad Sci U S A. 2000;97:14566–71. doi: 10.1073/pnas.97.26.14566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung A, Maier R, Vartanian JP, Bocharov G, Jung V, Fischer U, Meese E, Wain-Hobson S, Meyerhans A. Recombination: Multiply infected spleen cells in HIV patients. Nature. 2002;418:144. doi: 10.1038/418144a. [DOI] [PubMed] [Google Scholar]
- 26.Quinones-Mateu ME, Arts EJ. Recombination in HIV-1: Update and Implications. AIDS Rev. 1999;1:89–100. [Google Scholar]
- 27.McCutchan FE. Global epidemiology of HIV. J Med Virol. 2006;78(Suppl 1):S7–S12. doi: 10.1002/jmv.20599. [DOI] [PubMed] [Google Scholar]
- 28.Nora T, Charpentier C, Tenaillon O, Hoede C, Clavel F, Hance AJ. Contribution of recombination to the evolution of human immunodeficiency viruses expressing resistance to antiretroviral treatment. J Virol. 2007;81:7620–8. doi: 10.1128/JVI.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bender W, Chien Y-H, Chattopadhyay S, Vogt PK, Gardner MB, Davidson N. High-molecular-weight RNAs of AKR, NZB and wild mouse viruses and avian reticuloendotheliosis virus all have similar dimer structures. J Virol. 1978;25:888–896. doi: 10.1128/jvi.25.3.888-896.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kung HJ, Hu S, Bender W, Baily JM, Davidson N, Nicolson MO, McAllister RM. RD-114, baboon and wolly monkey viral RNAs compared in size and structure. Cell. 1976;7:609–620. doi: 10.1016/0092-8674(76)90211-7. [DOI] [PubMed] [Google Scholar]
- 31.Maisel J, Bender W, Hu S, Duesberg PH, Davidson N. Structure of 50 to 70S RNA from Moloney sarcoma viruses. J Virol. 1978;25:384–394. doi: 10.1128/jvi.25.1.384-394.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murti KG, Bondurant M, Tereba A. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. Journal of Virology. 1981;37:411–419. doi: 10.1128/jvi.37.1.411-419.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. Journal of Virology. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oritz-Conde BA, Hughes SH. Studies of the genomic RNA of leukosis viruses: implications for RNA dimerization. J Virol. 1999;73:7165–7174. doi: 10.1128/jvi.73.9.7165-7174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnoff R, Linial M. Specificity of retroviral RNA packaging. J Virol. 1991;65:71–80. doi: 10.1128/jvi.65.1.71-80.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnoff R, Hajjar AM, Linial ML. Avian retroviral RNA encapsidation: Reexamination of functional 5′ RNA sequences and the role of nucleocapsid Cys-His motifs. J Virol. 1993;67:178–188. doi: 10.1128/jvi.67.1.178-188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banks JD, Yeo A, Green K, Cepeda F, Linial ML. A minimal avian retroviral packaging sequence has a complex structure. J Virol. 1998;72:6190–6194. doi: 10.1128/jvi.72.7.6190-6194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doria-Rose NA, Vogt VM. In vivo selection of Rous sarcoma virus mutants with randomized sequences in packaging signals. J Virol. 1998;72:8073–8082. doi: 10.1128/jvi.72.10.8073-8082.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banks JD, Kealoha BO, Linial ML. An MΨ containing heterologous RNA, but not env mRNA, is efficiently packaged into avian retroviral particles. J Virol. 1999;73:8926–8933. doi: 10.1128/jvi.73.11.8926-8933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banks JD, Linial ML. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. J Virol. 2000;74:456–464. doi: 10.1128/jvi.74.1.456-464.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adam MA, Miller AD. Identification of a signal in a murine retrovirus that is sufficient for packaging of nonretroviral RNA into virions. Journal of Virology. 1988;62:3802–3806. doi: 10.1128/jvi.62.10.3802-3806.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mougel M, Barklis E. A role for two hairpin structures as a core RNA encapsidation signal in murine leukemia virus virions. J Virol. 1997;71:8061–8065. doi: 10.1128/jvi.71.10.8061-8065.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lever AML, Göttlinger HG, Haseltine WA, Sodroski JG. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. Journal of Virology. 1989;63:4085–4087. doi: 10.1128/jvi.63.9.4085-4087.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aldovini A, Young RA. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. Journal of Virology. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clavel F, Orenstein JM. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. Journal of Virology. 1990;64:5230–5234. doi: 10.1128/jvi.64.10.5230-5234.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poznansky M, Lever AML, Bergeron L, Haseltine W, Sodroski J. Gene transfer into human lymphocytes by a defective human immunodeficiency virus type 1 vector. Journal of Virology. 1991;65:532–536. doi: 10.1128/jvi.65.1.532-536.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clever JL, Parslow TG. Mutant Human Immunodeficiency Virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McBride MS, Panganiban AT. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McBride MS, Panganiban AT. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71:2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harrison GP, Miele G, Hunter E, Lever AML. Functional analysis of the core human immunodeficiency virus type 1 packaging signal in a permissive cell line. J Virol. 1998;72:5886–5896. doi: 10.1128/jvi.72.7.5886-5896.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim HJ, Lee K, O’Rear JJ. A short sequence upstream of the 5′ major splice site is important for encapsidation of HIV-1 genomic RNA. Virology. 1994;198:336–340. doi: 10.1006/viro.1994.1037. [DOI] [PubMed] [Google Scholar]
- 52.Laughrea M, Jette L, Mak J, Kleiman L, Liang C, Wainberg MA. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J Virol. 1997;71:3397–3406. doi: 10.1128/jvi.71.5.3397-3406.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laughrea M, Shen N, Jette L, Wainberg MA. Variant Effects of Non-Native Kissing-Loop Hairpin Palindromes on HIV Replication and HIV RNA Dimerization: Role of Stem-Loop B in HIV Replication and HIV RNA Dimerization. Biochemistry. 1999;38:226–234. doi: 10.1021/bi981728j. [DOI] [PubMed] [Google Scholar]
- 54.McBride MS, Schwartz MD, Panganiban AT. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Helga-Maria C, Hammarskjold M-L, Rekosh D. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type-1 genomic RNA. J Virol. 1999;73:4127–4135. doi: 10.1128/jvi.73.5.4127-4135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vicenzi E, Dimitrov DS, Engelman A, Migone TS, Purcell DF, Leonard J, Englund G, Martin MA. An integration-defective U5 deletion mutant of human immunodeficiency virus type 1 reverts by eliminating additional long terminal repeat sequences. J Virol. 1994;68:7879–7890. doi: 10.1128/jvi.68.12.7879-7890.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clever JL, Miranda JD, Parslow TG. RNA structure and packaging signals in the 5′ leader region of the human immunodeficiency virus type 1 genome. J Virol. 2002;76:12381–12387. doi: 10.1128/JVI.76.23.12381-12387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Russell RS, Hu J, Laughrea M, Wainberg MA, Liang C. Deficient dimerization of human immunodeficiency virus type 1 RNA caused by mutations of the U5 RNA sequences. Virology. 2002;303:152–163. doi: 10.1006/viro.2002.1592. [DOI] [PubMed] [Google Scholar]
- 59.Luban J, Goff SP. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. Journal of Virology. 1994;68:3784–3793. doi: 10.1128/jvi.68.6.3784-3793.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buchschacher JGL, Panganiban AT. Human immunodeficiency virus vectors for inducible expresson of foreign genes. J Virol. 1992;66:2731–2739. doi: 10.1128/jvi.66.5.2731-2739.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parolin C, Dorfman t, Palu G, Gottlinger HG, Sodroski J. Analysis in human immunodeficiency virus type 1 vectors of cis-acting sequences that affect gene transfer into human lymphocytes. J Virol. 1994;68:3888–3895. doi: 10.1128/jvi.68.6.3888-3895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paillart JC, Dettenhofer M, Yu X-F, Ehresmann C, Ehresmann B, Marquet R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J Biol Chem. 2004;279:48397–48403. doi: 10.1074/jbc.M408294200. [DOI] [PubMed] [Google Scholar]
- 63.Kuiken CL, Foley B, Freed EO, Hahn B, Marx P, McCutchan F, Mellors J, Wolinksy S, Korber B Group, T. B. a. B. HIV Sequence Compendium 2002. Los Alamos National Laboratory; 2002. LA-UR number 03–3564. [Google Scholar]
- 64.Mann R, Baltimore D. Varying the position of a retrovirus packaging sequence results in the encapsidation of both unspliced and spliced RNA. J Virol. 1985;54:401–407. doi: 10.1128/jvi.54.2.401-407.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prats AC, Roy C, Wang P, Erard M, Housset V, Gabus C, Paoletti C, Darlix JL. Cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. Journal of Virology. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.D’Souza V, Summers MF. Structural basis for packaging the dimeric genome of Moloney Murine Leukaemia Virus. Nature. 2004;431:586–590. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- 67.Miyazaki Y, Garcia E, King SR, Iyalla K, Loeliger K, Starck P, Syed S, Telesnitsky A, Summers MF. An RNA structural switch regulates diploid genome packaging by Moloney Murine Leukemia Virus. J Mol Biol. 2010;396:141–152. doi: 10.1016/j.jmb.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gherghe C, Lombo T, Leonard CW, Datta SA, Bess JW, Jr, Gorelick RJ, Rein A, Weeks KM. Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1006897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hayashi T, Shioda T, Iwakura Y, Shibuta H. RNA packaging signal of human immunodeficiency virus type 1. Virology. 1992;188:590–599. doi: 10.1016/0042-6822(92)90513-o. [DOI] [PubMed] [Google Scholar]
- 70.Hayashi T, Ueno Y, Okamoto T. Elucidation of a conserved RNA stem-loop structure in the packaging signal of human immunodeficiency virus type 1. FEBS. 1993;327:213–218. doi: 10.1016/0014-5793(93)80172-q. [DOI] [PubMed] [Google Scholar]
- 71.Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J. RNA interactions in the 5′ region of the HIV-1 genome. J Mol Biol. 2004;336:369–379. doi: 10.1016/j.jmb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 72.Harrison GP, Lever AML. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. Journal of Virology. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baudin F, Marquet R, Isel C, Darlix JL, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. Journal of Molecular Biology. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 74.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodefieiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clever J, Sassetti C, Parslow TG. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of Human Immunodeficiency Virus Type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Berkowitz RD, Ohagen A, Hoglund S, Goff SP. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol. 1995;69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dupraz P, Spahr PF. Specificity of Rous sarcoma virus nucleocapsid protein in genomic RNA packaging. Journal of Virology. 1992;66:4662–4670. doi: 10.1128/jvi.66.8.4662-4670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rizvi TA, Panganiban AT. Simian immunodeficiency virus RNA is efficiently encapsidate by human immunodeficiency virus type 1 particles. J Virol. 1993;67:2681–2688. doi: 10.1128/jvi.67.5.2681-2688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yin PD, Hu W-S. RNAs from genetically distinct retroviruses can copackage and exchange genetic information in vivo. J Virol. 1997;71:6237–6242. doi: 10.1128/jvi.71.8.6237-6242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kaye JF, Lever AM. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of gag in RNA encapsidation. Journal of virology. 1998;72:5877–5885. doi: 10.1128/jvi.72.7.5877-5885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Certo JL, Kabdulov TO, Paulson ML, Anderson JA, Hu W-S. The nucleocapsid domain is responsible for the ability of Spleen Necrosis Virus (SNV) Gag polyprotein to package SNV and Murine Leukemia Virus RNA. J Virol. 1999;73:9170–9177. doi: 10.1128/jvi.73.11.9170-9177.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fu W, Hu W-S. Functional replacement of nucleocapsid flanking regions by heterologous counterparts with divergent primary sequences: Effects of chimeric nucleocapsid on the retroviral replication cycle. J Virol. 2003;77:754–761. doi: 10.1128/JVI.77.1.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Poon DTK, Li g, Aldovini A. Nucleocapsid and matrix protein contributions to selective Human Immunodeficiency Virus Type 1 genomic RNA packaging. J Virol. 1998;72:1983–1993. doi: 10.1128/jvi.72.3.1983-1993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Henderson LE, Copeland TD, Sowder RC, Smythers GW, Oroslzan S. Primary structure of the low molecular weight nucleic acid-binding proteins of murine leukemia viruses. The Journal of Biological Chemistry. 1981;256:8400–8406. [PubMed] [Google Scholar]
- 86.Berg JM. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986;232:485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- 87.Schiff LA, Nibert ML, Fields BN. Characterization of a zinc blotting technique: Evidence that a retroviral gag protein binds zinc. Proceedings of the National Academy of Sciences USA. 1988;85:4195–4199. doi: 10.1073/pnas.85.12.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Smith LM, Jentoft JE. Conserved cysteine and histidine residues in avian myeloblastosis virus nucleocapsid protein pp12 are not zinc binding ligands. Biophysical Journal. 1988;53:295a. doi: 10.1073/pnas.85.19.7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jentoft JE, Smith LM, Fu X, Johnson M, Leis J. Conserved cysteine and histidine residues of the avian myeloblastosis virus nucleocapsid protein are essential for viral replication but are not “zinc-binding fingers”. Proceedings of the National Academy of Sciences USA. 1988;85:7094–7098. doi: 10.1073/pnas.85.19.7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gorelick RJ, Henderson LE, Hanser JP, Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: Evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc Natl Acad Sci USA. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Green LM, Berg JM. A retroviral Cys-Xaa2-Cys-Xaa4-His-Xaa4-Cys peptide binds metal ions: Spectoscopic studies and a proposed three-dimensional structure. Proceedings of the National Academy of Sciences USA. 1989;86:4047–4051. doi: 10.1073/pnas.86.11.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.South TL, Kim B, Summers MF. 113Cd NMR studies of a 1:1 Cd adduct with an 18-residue zinc finger peptide from HIV-1 nucleic acid binding protein, p7. Journal of the American Chemical Society. 1989;111:395–396. [Google Scholar]
- 93.South TL, Blake PR, Sowder RC, III, Arthur LO, Henderson LE, Summers MF. The nucleocapsid protein isolated from HIV-1 particles binds zinc and forms retroviral-type zinc fingers. Biochemistry. 1990;29:7786. doi: 10.1021/bi00486a002. [DOI] [PubMed] [Google Scholar]
- 94.Fitzgerald DW, Coleman JE. Physicochemical properties of cloned nucleocapsid protein from HIV. Interactions with metal ions. Biochemistry. 1991;30:5195–5201. doi: 10.1021/bi00235a012. [DOI] [PubMed] [Google Scholar]
- 95.Chance MR, Sagi I, Wirt MD, Frisbie SM, Scheuring E, Chen E, Bess JW, Jr, Henderson LE, Arthur LO, South TL, Perez-Alvarado G, Summers MF. Extended x-ray absorption fine structure studies of a retrovirus: Equine infectious anemia virus cysteine arrays are coordinated to zinc. Proceedings of the National Academy of Sciences USA. 1992;89:10041–10045. doi: 10.1073/pnas.89.21.10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Summers MF, Henderson LE, Chance MR, Bess JWJ, South TL, Blake PR, Sagi I, Perez-Alvarado G, Sowder RCI, Hare DR, Arthur LO. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Science. 1992;1:563–574. doi: 10.1002/pro.5560010502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Méric C, Gouilloud E, Spahr PF. Mutations in Rous sarcoma virus nucleocapsid protein p12 (NC): deletions of Cys-His Boxes. Journal of Virology. 1988;62:3328–3333. doi: 10.1128/jvi.62.9.3328-3333.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Méric C, Goff SP. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. Journal of Virology. 1989;63:1558–1568. doi: 10.1128/jvi.63.4.1558-1568.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gorelick RJ, Nigida J, Stephen M, Bess JW, Arthur LO, Henderson LE, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. Journal of Virology. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rice WG, Schaeffer CA, Harten B, Villinger F, South TL, Summers MF, Henderson LE, Bess JW, Jr, Arthur LO, McDougal JS, Orloff SL, Mendeleyev J, Kun E. Inhibition of HIV-1 infectivity by zinc-ejecting aromatic C-nitroso compounds. Nature. 1993;361:473–475. doi: 10.1038/361473a0. [DOI] [PubMed] [Google Scholar]
- 101.Rice WG, Supko JG, Malspeis L, Buckheit JRW, Clanton D, Bu M, Grahm L, Schaeffer CA, Turpin JA, Domagala J, Gogliotti R, Bader JP, Halliday SM, Coren L, Sowder RC, II, Arthur LO, Henderson LE. Inhibitors of HIV nucleocapsid protein zinc fingers as candidates for the treatment of AIDS. Science. 1995;270:1194–1197. doi: 10.1126/science.270.5239.1194. [DOI] [PubMed] [Google Scholar]
- 102.Rice WG, Turpin JA, Clanton D, Buckheit RW, Jr, Summers MF, McDonnell N, De Guzman RN, Covell DG, Wallqvist A, Zalkow L, Bader JP, Haugwitz RD, Sausville EA. Azodicarbonamide inhibits HIV-1 replication by targeting the nucleocapsid protein. Nature Medicine. 1997;3:341–345. doi: 10.1038/nm0397-341. [DOI] [PubMed] [Google Scholar]
- 103.McDonnell NB, De Guzman RN, Rice WG, Turpin JA, Summers MF. Zinc ejection as a new rationale for the use of cystamine and related dislufide-containing antiviral agents in the treatment of AIDS. J Med Chem. 1997;40:1969–1976. doi: 10.1021/jm970147+. [DOI] [PubMed] [Google Scholar]
- 104.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biology. 2008;6:883–899. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fu W, Gorelick RJ, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Feng Y-X, Copeland TD, Henderson LE, Gorelick RJ, Bosche WJ, Levin JG, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fu W, Ortiz-Conde BA, Gorelick RJ, Hughes SH, Rein A. Placement of tRNA primer on the primer-binding site requires pol gene expression in avian but not murine retroviruses. J Virol. 1997;71:6940–6946. doi: 10.1128/jvi.71.9.6940-6946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Feng Y-X, Campbell S, Harvin D, Ehresmann B, Ehresmann C, Rein A. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: Possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J Virol. 1999;73:4251–4256. doi: 10.1128/jvi.73.5.4251-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu W, Henderson LE, Copeland TD, Gorelick RJ, Bosche WJ, Rein A, Levin JG. Human immunodeficiency virus type 1 nucleocapsid protein reduces reverse transcriptase pausing at a secondary structure near the murine leukemia virus polypurine tract. J Virol. 1996;70:7132–7142. doi: 10.1128/jvi.70.10.7132-7142.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tanchou V, Gabus C, Rogemond V, Darlix J-L. Formation of stable and functional HIV-1 nucleoprotein complexes in vitro. J Mol Biol. 1995;252:563–571. doi: 10.1006/jmbi.1995.0520. [DOI] [PubMed] [Google Scholar]
- 111.Darlix J-L, Vincent C, Gabus H, de Rocquigny H, Roques B. Trans-activation of the 5′ to 3′ viral DNA strand transfer by nucleocapsid protein during reverse transcription of HIV-1. C R Acad Sci. 1993;361:763–771. [PubMed] [Google Scholar]
- 112.Allain B, Lapadat-Tapolsky M, Berlioz C, Darlix J-L. Transactivation of the minus-strand DNA transfer by nucleocapsid protein during reverse transcription of the retroviral genome. EMBO J. 1994;13:973–981. doi: 10.1002/j.1460-2075.1994.tb06342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Peliska JA, Balasubramanian S, Giedroc DP, Benkovic SJ. Recombinant HIV-1 nucleocapsid protein accelerates HIV-1 reverse transcriptase catalyzed DNA strand transfer reactions and modulates RNaseH activity. Biochemistry. 1994;33:13817–13823. doi: 10.1021/bi00250a036. [DOI] [PubMed] [Google Scholar]
- 114.Kim JK, Palaniappan C, Wu W, Fay PJ, Bambara RA. Evidence for a unique mechanism of strand transfer from the transactivation response region of HIV-1. J Biol Chem. 1997;272:16769–16777. doi: 10.1074/jbc.272.27.16769. [DOI] [PubMed] [Google Scholar]
- 115.Tsuchihashi Z, Brown PO. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–86. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 117.Darlix JL, Lapadat-Tapolsky M, de Rocquigny H, Roques BP. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. Journal of Molecular Biology. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- 118.Johnson PE, Turner RB, Wu ZR, Hairston L, Guo J, Levin JG, Summers MF. A mechanism for (+) strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription. Biochemistry. 2000;39:9084–9091. doi: 10.1021/bi000841i. [DOI] [PubMed] [Google Scholar]
- 119.Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc Natl Acad Sci U S A. 2001;98:6121–6. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Summers MF, South TL, Kim B, Hare DR. High-resolution structure of an HIV zinc fingerlike domain via a new NMR-based distance geometry approach. Biochemistry. 1990;29:329–340. doi: 10.1021/bi00454a005. [DOI] [PubMed] [Google Scholar]
- 121.South TL, Blake PR, Hare DR, Summers MF. C-terminal retroviral-type zinc finger domain from the HIV-1 nucleocapsid protein is structurally similar to the N-terminal zinc finger domain. Biochemistry. 1991;30:6342–6349. doi: 10.1021/bi00239a036. [DOI] [PubMed] [Google Scholar]
- 122.Klein D, Johnson PE, Zollars ES, De Guzman RN, Summers MF. The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle. Biochemistry. 2000;39:1604–1612. doi: 10.1021/bi9922493. [DOI] [PubMed] [Google Scholar]
- 123.Bertola F, Manigand C, Picard P, Belghazi M, Precigoux G. Human T-lymphotrophic virus type I nucleocapsid protein NCp15: Structural study and stability of the N-terminal zinc-finger. Biochem J. 2000;352:293–300. [PMC free article] [PubMed] [Google Scholar]
- 124.Mély Y, Piémont E, Sorinas-Jimeno M, de Rocquigny H, Julian N, Morellet N, Roques BP, Gérard D. Structural and dynamic characterization of the aromatic amino acids of the human immunodeficiency virus type 1 nucleocapsid protein zinc fingers and their involvement in heterologous tRNAPhe binding: A steady-state and time resolved flourescence study. Biophysical Journal. 1993;65:1513–1522. doi: 10.1016/S0006-3495(93)81222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Déméné H, Jullian N, Morellet N, de Rocquigny H, Cornille F, Maigret B, Roques BP. Three-dimensional 1H NMR structure of the nucleocapsid protein NCp10 of Moloney murine leukemia virus. J Biomol NMR. 1994;4:153–170. doi: 10.1007/BF00175244. [DOI] [PubMed] [Google Scholar]
- 126.Morellet N, Jullian N, de Rocquigny H, Maigret B, Darlix JL, Roques BP. Determination of the structure of the nucleocapsid protein NCp7 from the human immunodeficiency virus type 1 by 1H NMR. The EMBO Journal. 1992;11:3059–3065. doi: 10.1002/j.1460-2075.1992.tb05377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Morellet N, Meudal H, Bouaziz S, Roques BP. Structure of the zinc finger domain encompassing residues 13–51 of the nucleocapsid protein from simian immunodeficiency virus. Biochem J. 2006;393:725–32. doi: 10.1042/BJ20051203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kodera Y, Sato K, Tsukahara T, Ksmatsu H, Maeda T, Kohno T. High-resolution solution NMR structure of the minimal active domain of the human immunodeficiency virus type-2 nucleocapsid protein. Biochemistry. 1998;37:17704–17713. doi: 10.1021/bi981818o. [DOI] [PubMed] [Google Scholar]
- 129.Gao Y, Boyd J, Pielak GJ, Williams RJP. Proton Nuclear Magnetic Resonance as a Probe of Differences in Structure between the C102T and F82S, C102T Variants of Iso-1-cytochrome c from the Yeast Saccharomyces cerevisiae. Biochemistry. 1991;30:7033–7040. doi: 10.1021/bi00242a032. [DOI] [PubMed] [Google Scholar]
- 130.South TL, Summers MF. Zinc- and sequence-dependent binding to nucleic acids by the N-terminal zinc finger of the HIV-1 nucleocapsid protein: NMR structure of the complex with the Psi-site analog, dACGCC. Protein Science. 1993;2:3–19. doi: 10.1002/pro.5560020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.De Guzman RN, Turner RB, Summers MF. Protein-RNA recognition. Biopolymers. 1999;48:181–195. doi: 10.1002/(SICI)1097-0282(1998)48:2<181::AID-BIP7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 132.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 133.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor K, Wu Z-R, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the Ψ-RNA packaging signal. J Mol Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 134.Morellet N, Demene H, Teilleux V, Huynh-Dinh T, de Rocquigny H, Fournie-Zaluski M-C, Roques BP. Structure of the complex between the HIV-1 nucleocapsid protein NCp7 and the single-stranded pentanucleotide d(ACGCC) J Mol Biol. 1998;283:419–434. doi: 10.1006/jmbi.1998.2098. [DOI] [PubMed] [Google Scholar]
- 135.Zhou J, Bean RL, Vogt VM, Summers MF. Solution structure of the Rous sarcoma virus nucleocapsid protein:uY RNA packaging signal complex. J Mol Biol. 2006;365:453–467. doi: 10.1016/j.jmb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dey A, York D, Smalls-Mantey A, Summers MF. Composition and sequence dependent binding of RNA to the nucleocapsid protein of Moloney Murine Leukemia Virus. Biochemistry. 2005;44:3735–3744. doi: 10.1021/bi047639q. [DOI] [PubMed] [Google Scholar]
- 137.Spriggs S, Garyu L, Connor R, Summers MF. Potential intra- and intermolecular interactions involving the Unique-5′ Region of the HIV-1 5′-UTR. Biochemistry. 2008;46:13064–13073. doi: 10.1021/bi8014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pérez-Alvarado GC, Miles C, Michelsen JW, Louis HA, Winge DR, Beckerle MC, Summers MF. Structure of the carboxy-terminal LIM domain from the cysteine rich protein CRP. Nature Struct Biol. 1994;1:388–398. doi: 10.1038/nsb0694-388. [DOI] [PubMed] [Google Scholar]
- 139.Pérez-Alvarado GC, Kosa JL, Louis HA, Beckerle MC, Winge DR, Summers MF. Structure of the cysteine-rich intestinal protein, CRIP. J Mol Biol. 1996;257:153–174. doi: 10.1006/jmbi.1996.0153. [DOI] [PubMed] [Google Scholar]
- 140.Lee BM, De Guzman RN, Turner BG, Tjandra N, Summers MF. Dynamical behavior of the HIV-1 nucleocapsid protein. J Mol Biol. 1998;279:633–649. doi: 10.1006/jmbi.1998.1766. [DOI] [PubMed] [Google Scholar]
- 141.Demirov D, Freed EO. Retrovirus budding. Virus Research. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 142.Ono A, Freed EO. Role of lipid rafts in virus replication. Adv Virus Res. 2005;64:311–358. doi: 10.1016/S0065-3527(05)64010-9. [DOI] [PubMed] [Google Scholar]
- 143.Ono A, Orenstein JM, Freed EO. Role of the Gag Matrix Domain in Targeting Human Immunodeficiency Virus Type 1 assembly. Journal of Virology. 2000;74:2855–2866. doi: 10.1128/jvi.74.6.2855-2866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Waheed AA, Freed EO. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 2009;143:162–76. doi: 10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nash MA, Meyer MK, Decker GL, Arlinghaus RB. A subset of Pr65gag is nucleus associated in Murine Leukemia Virus-infected cells. J Virol. 1993;67:1350–1356. doi: 10.1128/jvi.67.3.1350-1356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dupont S, Sharova N, DeHoratius C, Virbasius CMA, Zhu X, Bukrinskaya AG, Stevenson M, Green MR. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature. 1999;402:681–685. doi: 10.1038/45272. [DOI] [PubMed] [Google Scholar]
- 147.Bukrinsky MI, Haggerty S, Dempsey MP, Sharova N, Adzhubei A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bukrinsky MI, Sharova N, Dempsey MP, Stanwick TL, Bukrinskaya AG, Haggerty S, Stevenson M. Active Nuclear Import of Human Immunodeficiency Virus Type 1 Preintegration Complexes. Proceedings of the National Academy of Sciences USA. 1992;89:6580–6584. doi: 10.1073/pnas.89.14.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bukrinsky MI, Sharova N, McDonald TL, Pushkarskaya T, Tarpley WG, Stevenson M. Association of Integrase, Matrix, and Reverse Transcriptase Antigens of Human Immunodeficiency Virus Type 1 with Viral Nucleic Acids following Acute Infection. Proceedings of the National Academy of Sciences USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Haffar OK, Popov S, Dubrovsky L, Agostini I, Tang H, Pushkarsky T, Nadler SG, Bukrinsky M. Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex. J Mol Biol. 2000;2999:359–368. doi: 10.1006/jmbi.2000.3768. [DOI] [PubMed] [Google Scholar]
- 151.Scheifele LZ, Garbitt RA, Rhoads JD, Parent LJ. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc Natl Acad Sci USA. 2002;99:3944–3949. doi: 10.1073/pnas.062652199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Callahan EM, Wills JW. Link between genome packaging and rate of budding for Rous Sarcoma Virus. Virology. 2003;77:9388–9398. doi: 10.1128/JVI.77.17.9388-9398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Parent LJ, Cairns TM, Albert JA, Wilson CB, Wills JW, Craven RC. RNA dimerization defect in a Rous sarcoma virus matrix mutant. J Virol. 2000;74:164–172. doi: 10.1128/jvi.74.1.164-172.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Garbitt-Hirst R, Kenney SP, Parent LJ. Genetic evidence for a connection between Rous Sarcoma Virus Gag nuclear trafficking and genomic RNA packaging. J Virol. 2009;83:6790–6797. doi: 10.1128/JVI.00101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Moore MD, Nikolaitchik OA, Chen J, Hammarskjold ML, Rekosh D, Hu WS. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 2009;5:e1000627. doi: 10.1371/journal.ppat.1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Steeg CM, Vogt VM. RNA-Binding Properties of the Matrix Protein (p19gag) of Avian Sarcoma and Leukemia Viruses. J Virol. 1990;64:847–855. doi: 10.1128/jvi.64.2.847-855.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Leis JP, Scheible P, Smith RE. Correlation of RNA binding affinity of avian oncornavirus p19 proteins with the extent of processing of virus genome RNA in cells. Journal of virology. 1980;35:722–31. doi: 10.1128/jvi.35.3.722-731.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cai M, Huang Y, Craigie R, Clore GM. Structural basis of the association of HIV-1 matrix protein with DNA. PloS one. 2010;5:e15675. doi: 10.1371/journal.pone.0015675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lochrie MA, Waugh S, Pratt JDG, Clever J, Parslow TG, Polisky B. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 1997;25:2902–2910. doi: 10.1093/nar/25.14.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Purohit P, Dupont S, Stevenson M, Green MR. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA. 2001;7:576–84. doi: 10.1017/s1355838201002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Alfadhli A, Still A, Barklis E. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J Virol. 2009;83:12196–203. doi: 10.1128/JVI.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chukkapalli V, Oh SJ, Ono A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc Natl Acad Sci U S A. 2010;107:1600–5. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jones CP, Datta SA, Rein A, Rouzina I, Musier-Forsyth K. Matrix Domain Modulates HIV-1 Gag’s Nucleic Acid Chaperone Activity via Inositol Phosphate Binding. J Virol. 2011;85:1594–603. doi: 10.1128/JVI.01809-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mangel WF, Delius H, Duesberg PH. Structure and molecular weight of the 60–70S RNA and the 30–40S RNA of the Rous sarcoma virus. Proc Natl Acad Sci USA. 1974;71:4541–4545. doi: 10.1073/pnas.71.11.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Levin JG, Grimley PM, Ramseur JM, Berezesky IK. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J Virol. 1974;14:152–161. doi: 10.1128/jvi.14.1.152-161.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Song R, Kafaie J, Yang L, Laughrea M. HIV-1 viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization. J Mol Biol. 2007;371:1084–1096. doi: 10.1016/j.jmb.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 169.Sakuragi J-I, Shioda T, Panganiban AT. Duplication of the primary encapsidation and dimer linkage region of Human immunodeficiency virus type 1 RNA results in the appearance of monomeric RNA in virions. J Virol. 2001;75:2557–2565. doi: 10.1128/JVI.75.6.2557-2565.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sakuragi J-I, Ueda S, Iwamoto A, Shioda T. Possible role of dimerization in human immunodeficiency virus Type-1 genome RNA packaging. J Virol. 2003;77:4060–4069. doi: 10.1128/JVI.77.7.4060-4069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Berkhout B, Vastenhouw NL, Klasens BI, Huthoff H. Structural features in the HIV-1 repeat region facilitate strand transfer during reverse transcription. RNA. 2001;7:1097–114. doi: 10.1017/s1355838201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Clever JL, Wong ML, Parslow TG. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Paillart J-C, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc Natl Acad Sci USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Paillart JC, Berthoux L, Ottmann M, Darlix JL, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J Virol. 1996;70:8348–54. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Berkhout B, van Wamel JL. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–32. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chin MPS, Rhodes TD, Chen J, Fu W, Hu WS. Identification of a major restriction in HIV-1 intersubtype recombination. Proc Natl Acad Sci USA. 2005;102:9002–9007. doi: 10.1073/pnas.0502522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Moore MD, Fu W, Nikolaitchik O, Chen J, Ptak RG, Hu W-S. Dimer initiation signal of human immunodeficiency virus type 1: Its role in partner selection during RNA copackaging and its effects on recombination. J Virol. 2007;81:4002–4011. doi: 10.1128/JVI.02589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kharytonchyk SA, Kireyeva AI, Osipovich AB, Fomin IK. Evidence for preferential copackaging of Moloney murine leukemia virus genomic RNAs transcribed in the sam chromosomal site. Retrovirology. 2005;1:1–11. doi: 10.1186/1742-4690-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rasmussen SV, Pedersen FS. Co-localization of gammaretroviral RNAs at their transcription site favours co-packaging. The Journal of general virology. 2006;87:2279–89. doi: 10.1099/vir.0.81759-0. [DOI] [PubMed] [Google Scholar]
- 180.Flynn JA, King SR, Telesnitsky A. Nonrandom dimerization of murine leukemia virus genomic RNAs. J Virol. 2004;78:12129–12139. doi: 10.1128/JVI.78.22.12129-12139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Flynn JA, Telesnitsky A. Two distinct Moloney murine leukemia virus RNAs produced from a single locus dimerize at random. Virology. 2006;344:391–400. doi: 10.1016/j.virol.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rhodes T, Wargo H, Hu W-S. High rates of human immunodeficiency virus type 1 recombination: Near-random segregation of markers one kilobase apart in one round of viral replication. J Virol. 2003;77:11193–11200. doi: 10.1128/JVI.77.20.11193-11200.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Onafuwa A, An W, Robson ND, Telesnitsky A. Human immunodeficiency virus type 1 genetic recombination is more frequent than that of Moloney murine leukemia virus despite similar template switching rates. J Virol. 2003;77:4577–4587. doi: 10.1128/JVI.77.8.4577-4587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bennasser Y, Le SY, Yeung ML, Jeang KT. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology. 2004;1:43. doi: 10.1186/1742-4690-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Berkowitz RD, Hammarskjold ML, Helga-Maria C, Rekosh D, Goff SP. 5′ Regions of HIV-1 RNAs are not sufficient for encapsidation: Implications for the HIV-1 packaging signal. Virology. 1995;212:718–723. doi: 10.1006/viro.1995.1530. [DOI] [PubMed] [Google Scholar]
- 186.Kung H-J, Hu S, Bender W, Baily JM, Davidson N, Nicolson MO, McAllister RM. Structure, subunit composition, and molecular weight of RD-114 RNA. J Virol. 1975;16:397–411. doi: 10.1128/jvi.16.2.397-411.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bender W, Davidson N. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell. 1976;7:595–607. doi: 10.1016/0092-8674(76)90210-5. [DOI] [PubMed] [Google Scholar]
- 188.Höglund S, Öhagen A, Goncalves J, Panganiban AT, Gabuzda D. Ultrastructure of HIV-1 genomic RNA. Virology. 1997;233:271–279. doi: 10.1006/viro.1997.8585. [DOI] [PubMed] [Google Scholar]
- 189.Berkhout B. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 1992;20:27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Berkhout B, Klaver B, Das AT. A conserved hairpin structure predicted for the poly(A) signal of human and simian immunodeficiency viruses. Virology. 1995;207:276–81. doi: 10.1006/viro.1995.1077. [DOI] [PubMed] [Google Scholar]
- 191.Carroll R, Peterlin BM, Derse D. Inhibition of human immunodeficiency virus type 1 Tat activity by coexpression of heterologous trans activators. Journal of virology. 1992;66:2000–7. doi: 10.1128/jvi.66.4.2000-2007.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wimmer J, Fujinaga K, Taube R, Cujec TP, Zhu Y, Peng J, Price DH, Peterlin BM. Interactions between Tat and TAR and human immunodeficiency virus replication are facilitated by human cyclin T1 but not cyclins T2a or T2b. Virology. 1999;255:182–9. doi: 10.1006/viro.1998.9589. [DOI] [PubMed] [Google Scholar]
- 193.Bannwarth S, Gatignol A. HIV-1 TAR RNA: the target of molecular interactions between the virus and its host. Curr HIV Res. 2005;3:61–71. doi: 10.2174/1570162052772924. [DOI] [PubMed] [Google Scholar]
- 194.Brady J, Kashanchi F. Tat gets the “green” light on transcription initiation. Retrovirology. 2005;2:69. doi: 10.1186/1742-4690-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Das AT, Klaver B, Berkhout B. The 5′ and 3′ TAR elements of human immunodeficiency virus exert effects at several points in the virus life cycle. J Virol. 1998;72:9217–23. doi: 10.1128/jvi.72.11.9217-9223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Clever JL, Eckstein DA, Parslow TG. Genetic dissociation of the encapsidation and reverse transcription functions in the 5′ R region of human immunodeficiency virus type 1. J Virol. 1999;73:101–109. doi: 10.1128/jvi.73.1.101-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Verhoef K, Marzio G, Hillen W, Bujard H, Berkhout B. Strict control of human immunodeficiency virus type 1 replication by a genetic switch: Tet for Tat. J Virol. 2001;75:979–87. doi: 10.1128/JVI.75.2.979-987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Das AT, Harwig A, Vrolijk MM, Berkhout B. The TAR hairpin of human immunodeficiency virus Type 1 can be deleted when not required for Tat-mediated activation of transcription. J Virol. 2007;81:7742–7748. doi: 10.1128/JVI.00392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Berkhout B. Prog Nucl Acid Res and Mol Biol. Vol. 54. Academic Press, Inc; 1996. Structure and function of the human immunodeficiency virus leader RNA; pp. 1–34. [DOI] [PubMed] [Google Scholar]
- 200.Aboul-ela F, Karn J, Varani G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals prinicples of RNA recognition by Tat protein. J Mol Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 201.Hauber J, Malim MH, Cullen BR. Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. Journal of virology. 1989;63:1181–7. doi: 10.1128/jvi.63.3.1181-1187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Delling U, Reid LS, Barnett RW, Ma MY, Climie S, Sumner-Smith M, Sonenberg N. Conserved nucleotides in the TAR RNA stem of human immunodeficiency virus type 1 are critical for Tat binding and trans activation: model for TAR RNA tertiary structure. Journal of virology. 1992;66:3018–25. doi: 10.1128/jvi.66.5.3018-3025.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, Lowe AD, Singh M, Skinner MA. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. The EMBO journal. 1990;9:4145–53. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 205.Richter S, Ping YH, Rana TM. TAR RNA loop: a scaffold for the assembly of a regulatory switch in HIV replication. Proc Natl Acad Sci U S A. 2002;99:7928–33. doi: 10.1073/pnas.122119999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer) J Mol Biol. 1995;247:236–50. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- 207.Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 208.Goldschmidt V, Rigourd M, Ehresmann C, Le Grice SF, Ehresmann B, Marquet R. Direct and indirect contributions of RNA secondary structure elements to the initiation of HIV-1 reverse transcription. J Biol Chem. 2002;277:43233–42. doi: 10.1074/jbc.M205295200. [DOI] [PubMed] [Google Scholar]
- 209.Isel C, Westhof E, Massire C, Le Grice SF, Ehresmann B, Ehresmann C, Marquet R. Structural basis for the specificity of the initiation of HIV-1 reverse transcription. EMBO J. 1999;18:1038–48. doi: 10.1093/emboj/18.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Shen N, Jette L, Liang C, Wainberg MA, Laughrea M. Impact of human immunodeficiency virus type 1 RNA dimerization on viral infectivity and of stem-loop B on RNA dimerization and reverse transcription and dissociation of dimerization from packaging. J Virol. 2000;74:5729–35. doi: 10.1128/jvi.74.12.5729-5735.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Athavale SS, Ouyang W, McPike MP, Hudson BS, Borer PN. Effects of the nature and concentration of salt on the interaction of the HIV-1 nucleocapsid protein with SL3 RNA. Biochemistry. 2010;49:3525–33. doi: 10.1021/bi901279e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Berkhout B. The primer binding site on the RNA genome of human and simian immunodeficiency viruses is flanked by an upstream hairpin structure. Nucleic Acids Res. 1997;25:4013–7. doi: 10.1093/nar/25.20.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Beerens N, Groot F, Berkhout B. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J Biol Chem. 2001;276:31247–56. doi: 10.1074/jbc.M102441200. [DOI] [PubMed] [Google Scholar]
- 214.Jaeger L, Michel F, Westhof E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J Mol Biol. 1994;236:1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 215.Haddrick M, Lear AL, Cann AJ, Heaphy S. Evidence that a kissing loop structure facilitates genomic RNA dimerisation in HIV-1. J Mol Biol. 1996;259:58–68. doi: 10.1006/jmbi.1996.0301. [DOI] [PubMed] [Google Scholar]
- 216.Ye X, Kumar RA, Patel DJ. Molecular recognition in the bovine immunodeficiency virus tat peptide-TAR RNA complex. Chem Biol. 1995;2:827–840. doi: 10.1016/1074-5521(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 217.Pitt SW, Majumdar A, Serganov A, Patel DJ, Al-Hashimi HM. Argininamide binding arrests global motions in HIV-1 TAR RNA: comparison with Mg2+-induced conformational stabilization. J Mol Biol. 2004;338:7–16. doi: 10.1016/j.jmb.2004.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Puglisi JD, Chen L, Frankel AD, Williamson JR. Role of RNA structure in arginine recognition of TAR RNA. Proceedings of the National Academy of Sciences USA. 1993;90:3680–3684. doi: 10.1073/pnas.90.8.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Calnan BJ, Tidor B, Biancalana S, Hudson D, Frankel AD. Arginine-mediated RNA recognition: The arginine fork. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- 220.Zhang Q, Al-Hashimi HM. Domain-elongation NMR spectroscopy yields new insights into RNA dynamics and adaptive recognition. RNA. 2009;15:1941–8. doi: 10.1261/rna.1806909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Zhang Q, Sun X, Watt ED, Al-Hashimi HM. Resolving the motional modes that code for RNA adaptation. Science. 2006;311:653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- 222.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450:1263–1267. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 223.Bryson DI, Zhang W, Ray WK, Santos WL. Screening of a branched peptide library with HIV-1 TAR RNA. Mol Biosyst. 2009;5:1070–3. doi: 10.1039/b904304g. [DOI] [PubMed] [Google Scholar]
- 224.Duca M, Malnuit V, Barbault F, Benhida R. Design of novel RNA ligands that bind stem-bulge HIV-1 TAR RNA. Chem Commun (Camb) 2010;46:6162–4. doi: 10.1039/c0cc00645a. [DOI] [PubMed] [Google Scholar]
- 225.Das AT, Klaver B, Klasens BI, van Wamel JL, Berkhout B. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J Virol. 1997;71:2346–56. doi: 10.1128/jvi.71.3.2346-2356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Das AT, Klaver B, Berkhout B. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J Virol. 1999;73:81–91. doi: 10.1128/jvi.73.1.81-91.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Abbink TE, Ooms M, Haasnoot PC, Berkhout B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry. 2005;44:9058–66. doi: 10.1021/bi0502588. [DOI] [PubMed] [Google Scholar]
- 228.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. In vitro evidence for a long range pseudoknot in the 5′-untranslated and matrix coding regions of the HIV-1 genomic RNA. J Biol Chem. 2002;277:5995–6004. doi: 10.1074/jbc.M108972200. [DOI] [PubMed] [Google Scholar]
- 229.Zhang Z, Yu Q, Kang SM, Buescher J, Morrow CD. Preferential completion of human immunodeficiency virus type 1 proviruses initiated with tRNA3Lys rather than tRNA1,2Lys. J Virol. 1998;72:5464–71. doi: 10.1128/jvi.72.7.5464-5471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Beerens N, Berkhout B. The tRNA primer activation signal in the human immunodeficiency virus type 1 genome is important for initiation and processive elongation of reverse transcription. J Virol. 2002;76:2329–39. doi: 10.1128/jvi.76.5.2329-2339.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Isel C, Lanchy JM, Le Grice SF, Ehresmann C, Ehresmann B, Marquet R. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcription require the post-transcriptional modifications of primer tRNA3Lys. EMBO J. 1996;15:917–24. [PMC free article] [PubMed] [Google Scholar]
- 232.Damgaard CK, Dyhr-Mikkelsen H, Kjems J. Mapping the RNA binding sites for human immunodeficiency virus type-1 Gag and NC proteins within the complete HIV-1 and HIV-2 untranslated leader regions. Nucleic Acids Res. 1998;26:3667–3676. doi: 10.1093/nar/26.16.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nature Prot. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 234.Abbink TEM, Berkhout B. A novel long distance base-pairing interaction in Human Immunodeficiency Virus Type 1 RNA occludes the Gag start codon. J Biol Chem. 2003;278:11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- 235.Russell RS, Hu J, Beriault V, Mouland AJ, Kleiman L, Wainberg MA, Liang C. Sequences downstream of the 5′ splice donor site are required for both packaging and dimerization of human immunodeficiency virus type-1 RNA. J Virol. 2003;77:84–96. doi: 10.1128/JVI.77.1.84-96.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Paillart JC, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J Biol Chem. 1994;269:27486–27493. [PubMed] [Google Scholar]
- 237.Lawrence DC, Stover CC, Noznitsky J, Wu Z-R, Summers MF. Structure of the intact stem and bulge of HIV-1 Ψ-RNA stem loop SL1. J Mol Biol. 2003;326:529–542. doi: 10.1016/s0022-2836(02)01305-0. [DOI] [PubMed] [Google Scholar]
- 238.Mujeeb A, Clever JL, Billeci TM, James TL, Parslow TG. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nature Struct Biol. 1998;5:432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 239.Kieken F, Paquet F, Brule F, Paoletti J, Lancelot G. A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer. Nucleic Acids Res. 2006;34:343–52. doi: 10.1093/nar/gkj427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 241.Girard F, Barbault F, Gouyett C, Huynh-dinh T, Paoletti J, Lamcelot G. Dimer initiation sequence of HIV-1 Lai genomic RNA: NMR solution structure of the extended nucleus. J of Biomol Struct & Dyn. 1999;16:1145–1157. doi: 10.1080/07391102.1999.10508323. [DOI] [PubMed] [Google Scholar]
- 242.Mihailescu MR, Marino JP. A proton-coupled dynamic conformational switch in the HIV-1 dimerization initiation site kissing complex. Proc Natl Acad Sci USA. 2004;101:1189–1194. doi: 10.1073/pnas.0307966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ennifar E, Walter P, Ehresmann B, Ehresmann C, Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat Struct Biol. 2001;8:1064–1068. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 244.Ennifar E, Yusupov M, Walter P, Marquet R, Ehresmann B, Ehresmann C, Dumas P. The crystal structure of the dimerization initiation site of genomic HIV-1 RNA reveals an extended duplex with two adenine bulges. Structure. 1999;7:1439–1449. doi: 10.1016/s0969-2126(00)80033-7. [DOI] [PubMed] [Google Scholar]
- 245.Muriaux D, Fosse P, Paoletti J. A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. Biochemistry. 1996;35:5075–82. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- 246.Ulyanov NB, Mujeeb A, Du Z, Tonelli M, Parslow TG, James TL. NMR structure of the full-length linear dimer of stem-loop-1 RNA in the HIV-1 dimer initiation site. J Biol Chem. 2006;281:16168–77. doi: 10.1074/jbc.M601711200. [DOI] [PubMed] [Google Scholar]
- 247.Mujeeb A, Parslow TG, Zarrinpar A, Das C, James TL. NMR structure of the mature dimer initiation complex of HIV-1 genomic RNA. FEBS Letters. 1999;458:387–392. doi: 10.1016/s0014-5793(99)01183-7. [DOI] [PubMed] [Google Scholar]
- 248.Muriaux D, de Rocquigny H, Roques BP, Paoletti J. NCp7 activates HIV-1Lai RNA dimerization by converting a transient loop-loop complex into a stable dimer. J Biol Chem. 1996;271:33686–33692. doi: 10.1074/jbc.271.52.33686. [DOI] [PubMed] [Google Scholar]
- 249.Darlix J-L, Gabus C, Nugeyre M-T, Clavel F, Barre-Sinoussi F. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J Mol Biol. 1990;216:689–699. doi: 10.1016/0022-2836(90)90392-Y. [DOI] [PubMed] [Google Scholar]
- 250.Yuan Y, Kerwood DJ, Paoletti AC, Shubsda MF, Borer PN. Stem of SL1 RNA in HIV-1: structure and nucleocapsid protein binding for a 1 × 3 internal loop. Biochemistry. 2003;42:5259–69. doi: 10.1021/bi034084a. [DOI] [PubMed] [Google Scholar]
- 251.Sun X, Zhang Q, Al-Hashimi HM. Resolving fast and slow motions in the internal loop containing stem-loop 1 of HIV-1 that are modulated by Mg2+ binding: role in the kissing-duplex structural transition. Nucleic Acids Res. 2007;35:1698–713. doi: 10.1093/nar/gkm020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Laughrea M, Shen N, Jette L, Darlix JL, Kleiman L, Wainberg MA. Role of distal zinc finger of nucleocapsid protein in genomic RNA dimerization of human immunodeficiency virus type 1; no role for the palindrome crowning the R-U5 hairpin. Virology. 2001;281:109–16. doi: 10.1006/viro.2000.0778. [DOI] [PubMed] [Google Scholar]
- 253.Amarasinghe GK, De Guzman RN, Turner RB, Summers MF. NMR structure of stem loop SL2 of the HIV-1 Ψ-RNA packaging signal reveals a novel A-U-A base triple platform. J Mol Biol. 2000;299:145–156. doi: 10.1006/jmbi.2000.3710. [DOI] [PubMed] [Google Scholar]
- 254.Clever JL, Taplitz RA, Lochrie MA, Polisky B, Parslow TG. A heterologous, high-affinity RNA ligand for human immunodeficiency virus Gag protein has RNA packaging activity. J Virol. 2000;74:541–546. doi: 10.1128/jvi.74.1.541-546.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Pappalardo L, Kerwood DJ, Pelczer I, Borer PN. Three-dimensional folding of an RNA hairpin required for packaging HIV-1. J Mol Biol. 1998;282:801–818. doi: 10.1006/jmbi.1998.2046. [DOI] [PubMed] [Google Scholar]
- 256.Song R, Kafaie J, Laughrea M. Role of the 5′ TAR stem-loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA. Biochemistry. 2008 doi: 10.1021/bi7023173. in press. [DOI] [PubMed] [Google Scholar]
- 257.Nikolaitchik O, Rhodes TD, Ott D, Hu W-S. Effects of mutations in the Human Immunodeficiency Virus Type 1 gag gene on RNA packaging and recombination. J Virol. 2006;80:4691–4697. doi: 10.1128/JVI.80.10.4691-4697.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Abramovitz DL, Pyle AM. Remarkable morphological variability of a common RNA folding motif: the GNRA tetraloop-receptor interaction. J Mol Biol. 1997;266:493–506. doi: 10.1006/jmbi.1996.0810. [DOI] [PubMed] [Google Scholar]
- 259.Jucker FM, Heus HA, Yip PF, Moors EHM, Pardi A. A Network of heterogeneous hydrogen bonds in GNRA tetraloops. J Mol Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- 260.Kerwood DJ, Cavaluzzi MJ, Borer PN. Structure of SL4 RNA from the HIV-1 packaging signal. Biochemistry. 2001;40:14518–29. doi: 10.1021/bi0111909. [DOI] [PubMed] [Google Scholar]
- 261.Amarasinghe GK, Zhou J, Miskimon M, Chancellor KJ, McDonald JA, Matthews AG, Miller RA, Rouse MD, Summers MF. Stem-loop SL4 of the HIV-1Ψ-RNA packaging signal exhibits weak affinity for the nucleocapsid protein. Structural studies and implications for genome recognition. J Mol Biol. 2001;314:961–969. doi: 10.1006/jmbi.2000.5182. [DOI] [PubMed] [Google Scholar]
- 262.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–6. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Lu K, Miyazaki Y, Summers MF. Isotope labeling strategies for NMR studies of RNA. J Biomol NMR. 2009;46:113–125. doi: 10.1007/s10858-009-9375-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Yu ET, Hawkins A, Eaton J, Fabris D. MS3D structural elucidation of the HIV-1 packaging signal. Proc Natl Acad Sci USA. 2008;105:12248–12253. doi: 10.1073/pnas.0800509105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Baig TT, Strong CL, Lodmell JS, Lanchy JM. Regulation of primate lentiviral RNA dimerization by structural entrapment. Retrovirology. 2008;5:65. doi: 10.1186/1742-4690-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Berkhout B, Van Wamel JLB. The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA. 2000;6:282–295. doi: 10.1017/s1355838200991684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Huthoff H, Berkhout B. Two alternating structures of the HIV-1 leader RNA. RNA. 2001;7:143–157. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Berkhout B, Ooms M, Beerens N, Huthoff H, Southern E, Verhoef K. In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J Biol Chem. 2002;277:19967–19975. doi: 10.1074/jbc.M200950200. [DOI] [PubMed] [Google Scholar]
- 269.Ooms M, Huthoff H, Russell R, Liang C, Berkhout B. A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J Virol. 2004;78:10814–10819. doi: 10.1128/JVI.78.19.10814-10819.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Stoltzfus CM, Snyder PN. Structure of B77 sarcoma virus RNA: stabilization of RNA after packaging. J Virol. 1975;16:1161–1170. doi: 10.1128/jvi.16.5.1161-1170.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Ivanchenko S, Godinez WJ, Lampe M, Krausslich HG, Eils R, Rohr K, Brauchle C, Muller B, Lamb DC. Dynamics of HIV-1 assembly and release. PLoS pathogens. 2009;5:e1000652. doi: 10.1371/journal.ppat.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kemler I, Meehan A, Poeschla EM. Live-cell coimaging of the genomic RNAs and Gag proteins of two lentiviruses. Journal of virology. 2010;84:6352–66. doi: 10.1128/JVI.00363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nature cell biology. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Miyazaki Y, Irobalieva RN, Tolbert BS, Smalls-Manty A, Iyalla K, Loeliger K, D’Souza V, Khant H, Schmid MF, Garcia E, Telesnitsky A, Chiu W, Summers MF. Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. J Mol Biol. 2010;404:751–772. doi: 10.1016/j.jmb.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Badorrek CS, Gherghe CM, Weeks KM. Structure of an RNA switch that enforces stringent retroviral genomic RNA dimerization. Proc Natl Acad Sci USA. 2006;103:13640–13645. doi: 10.1073/pnas.0606156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Davidson A, Leeper TC, Athanassiou Z, Patora-Komisarska K, Karn J, Robinson JA, Varani G. Simultaneous recognition of HIV-1 TAR RNA bulge and loop sequences by cyclic peptide mimics of Tat protein. Proc Natl Acad Sci U S A. 2009;106:11931–6. doi: 10.1073/pnas.0900629106. [DOI] [PMC free article] [PubMed] [Google Scholar]