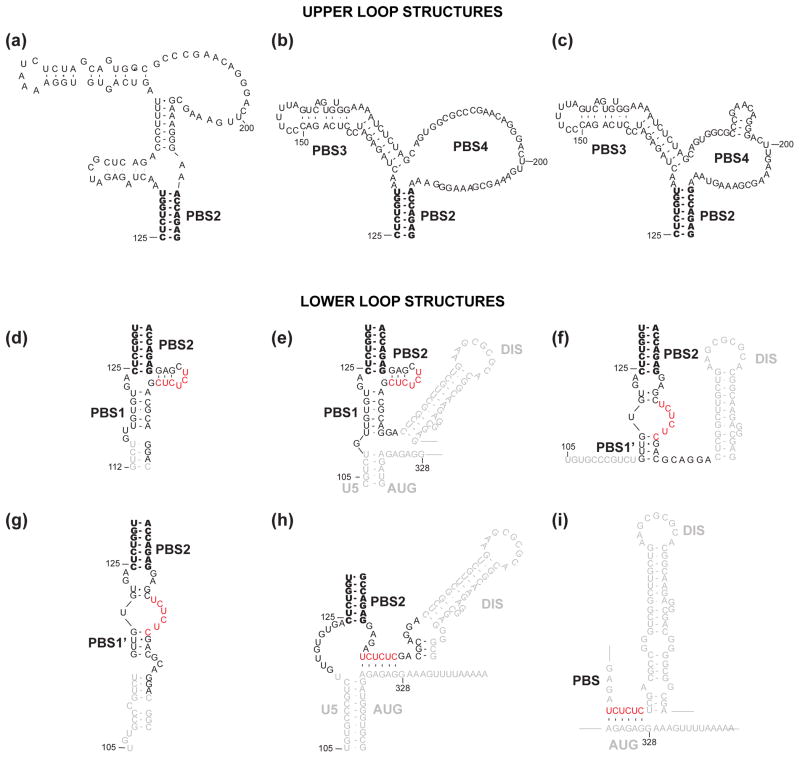
Figure 4.
Secondary structures predicted for the upper (a–c) and lower (d–i) loops of the HIV-1 PBS. Nearly all the models include a common base paired helix in the center of the PBS (PBS2, in bold). Upper loop predictions: (a) Berkhout and co-workers predicted a structure containing an apical hairpin ending in an A-rich loop199 for the HIV-1 HXB-2 RNA genome. (b) Damgaard et al. proposed that the important A-rich sequence is displayed in an internal loop232. (c) Wilkinson and co-workers further predicted a small hairpin structure in the PBS4 region, based on the SHAPE probing104 for the HIV-1 NL4-3 RNA genome. Lower loop predictions: (d,e) Kjems and co-workers predicted that the CU rich region (red) forms a small hairpin with the nucleotides right after the PBS2 stem loop71,232. (f,g) Berkhout and co-workers proposed that CU rich region forms a loop structure199,227. (h) Weeks and co-workers predicted that the CU rich region forms a long-range interaction with the AG rich linker between Ψ and AUG104,262, consistent with an earlier prediction by Lever and co-workers72 (i).