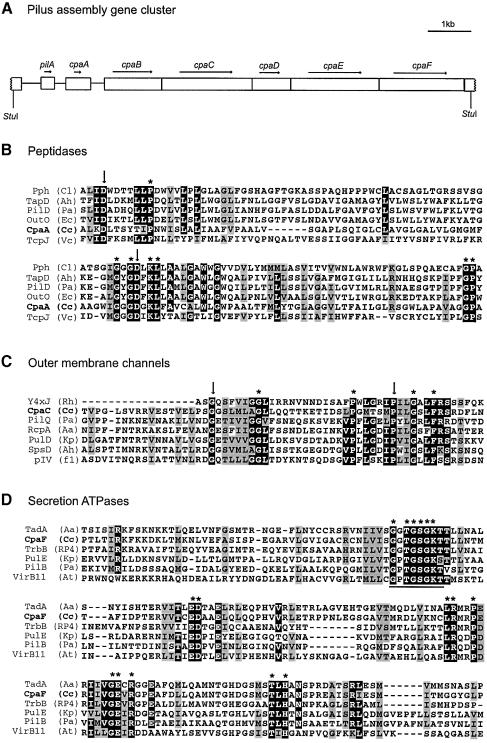
Fig. 3. Sequence analysis of a pilus assembly gene cluster. (A) Diagram of an 8 kb StuI fragment that complements transposon mutant Tn24-3. Seven open reading frames were identified on this fragment, including the pilA locus encoding the pilin subunit and three other genes with homology to known pilus assembly proteins. (B) Alignment of the deduced amino acid sequence of Caulobacter CpaA with several members of the prepilin peptidase family. Only the region of the protein that is thought to contain the active site of the peptidase has been aligned. Putative active site residues are marked with an arrow. Invariant residues are marked with an asterisk. The sequences used in this alignment are Chlorobium limicola plasmid pCL1, Pph (Cl; DDBJ/EMBL/GenBank accession No. U77780), Aeromonas hydrophila TapD (Ah; DDBJ/EMBL/GenBank accession No. U20255), P.aeruginosa PilD (Pa; DDBJ/EMBL/GenBank accession No. M32066), Erwinia caratovora OutO (Ec; DDBJ/EMBL/GenBank accession No. X70049), C.crescentus CpaA (Cc) and V.cholerae TcpJ (Vc; DDBJ/EMBL/GenBank accession No. M74708). (C) Caulobacter CpaC is similar to the PulD/pIV family of outer membrane channels. Only the most highly conserved region of this protein family has been aligned. Invariant residues are marked with an asterisk and two functionally important residues are indicated with an arrow. The sequences used in this alignment are Rhizobium sp. NGR234 Y4×J (Rh; DDBJ/EMBL/GenBank accession No. AE000106), C.crescentus CpaC (Cc), P.aeruginosa PilQ (Pa; DDBJ/EMBL/GenBank accession No. L13865), A.actinomycetemcomitans RcpA (Aa; DDBJ/EMBL/GenBank accession No. AF139249), Klebsiella pneumoniae PulD (Kp; DDBJ/EMBL/GenBank accession No. M32613), A.hydrophila SpsD (Ah; DDBJ/EMBL/GenBank accession No. L41682) and coliphage f1 pIV (f1; DDBJ/EMBL/GenBank accession No. V00606). (D) Caulobacter CpaF is a member of the TrbB/VirB11 family of secretion ATPases. Only the most highly conserved region, surrounding the Walker Box (underlined), has been aligned. Invariant residues are marked with an asterisk. The sequences used in this alignment are A.actinomycetemcomitans TadA (Aa; DDBJ/EMBL/GenBank accession No. AF152598), C.crescentus CpaF (Cc), plasmid RP4 TrbB (RP4; DDBJ/EMBL/GenBank accession No. M93696), K.pneumoniae PulE (Kp; DDBJ/EMBL/GenBank accession No. M32613), P.aeruginosa PilB (Pa; DDBJ/EMBL/GenBank accession No. M32066) and Agrobacterium tumefaciens plasmid pTiC58, VirB11 (At; DDBJ/EMBL/GenBank accession No. X53264).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.