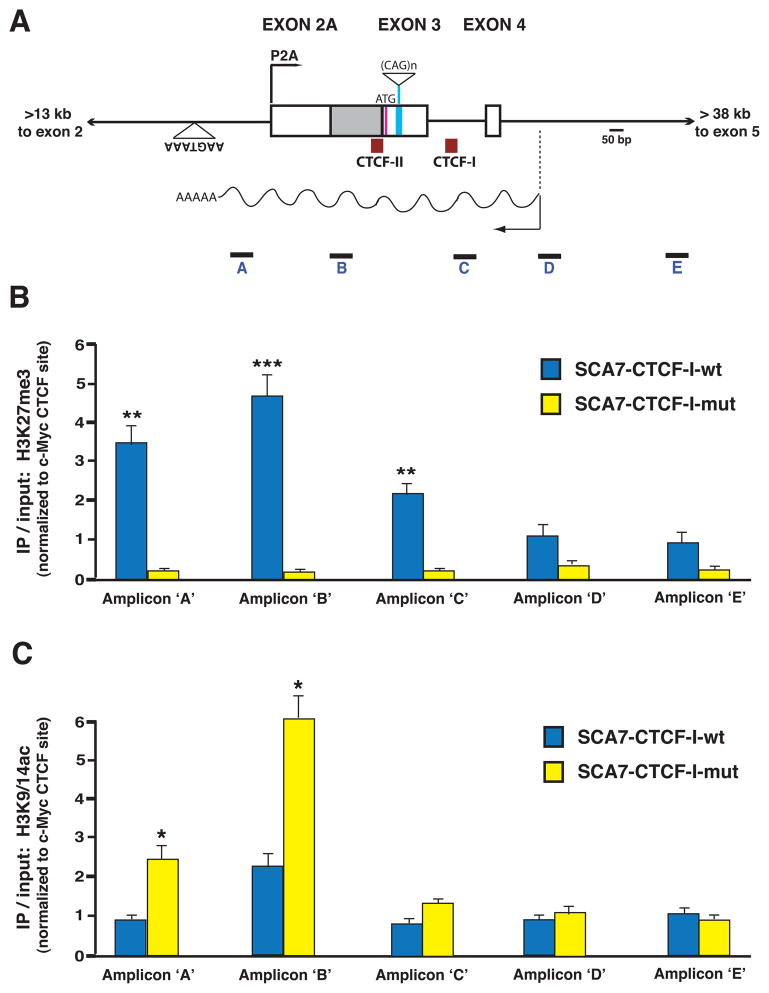
Figure 7. ChIP analysis of histone modification status at the ataxin-7 alternative promoter region in SCA7 transgenic mice.
A) Diagram of the genomic region of the human ataxin-7 gene, where the alternative promoter (P2A), first coding exon, CTCF binding sites, and antisense ncRNA, SCAANT1, are located. Boxes represent exons, while solid lines correspond to introns. The positions of the five amplicons employed in the ChIP analysis are indicated.
B–C) Results of ChIP analysis of histone marks in the cerebellum of SCA7 transgenic mice. ChIP was performed with antibodies against a repressive mark (H3K27me3) and activated transcription mark (H3K9/14Ac) on cerebellar DNAs isolated from six-month old SCA7-CTCF-I-wt and SCA7-CTCF-I-mut mice. B) In SCA7-CTCF-I-wt mice that express minimal amounts of ataxin-7 sense RNA from P2A, there was significant enrichment of H3K27me3 extending from the alternative promoter region into intron 3 of the ataxin-7 gene (p < .01 to .001, ANOVA). C) This pattern was reversed in SCA7-CTCF-I-mut mice that display robust activation of the P2A promoter, as significant enrichment of the H3K9/14Ac mark was present in the ataxin-7 sense P2A region and initial transcript region (p < 0.05, ANOVA). Results represent three independent experiments quantifying histone antibody ChIP normalized to the c-Myc promoter region. Error bars = s.e.m.