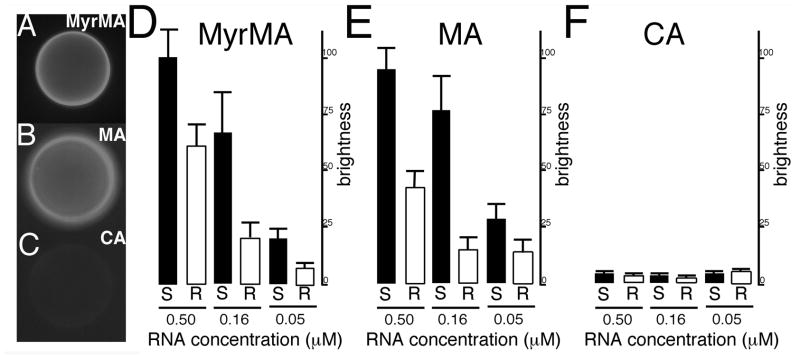
Figure 1. Fluorescent RNA bead binding assay.
In panels (A–C), binding assays were performed with 0.16 μM FITC-Sel25 RNA and beads coated with (A) MyrMA, (B) MA or (C) CA proteins. After binding and washing steps, beads with bound RNA were imaged by fluorescence microscopy under identical gain and exposure settings, and photographed: note that bead brightness is indicative of RNA binding. In panels (D–F) levels of binding were quantitated for beads coated with the indicated proteins and FITC-Sel25 RNA (black bars) or FITC-Ran25 RNA (white bars) at the indicated 0.5, 0.16, and 0.05 μM RNA concentrations. For quantitation, background-subtracted bead brightness values were calculated, averaged from multiple beads for each incubation, and normalized to MyrMA plus FITC-Sel25 RNA (0.5 μM) signals. For capsid beads, averages derive from four readings each; for matrix beads, averages derive from 12 separate beads collected from experiments on three separate dates.