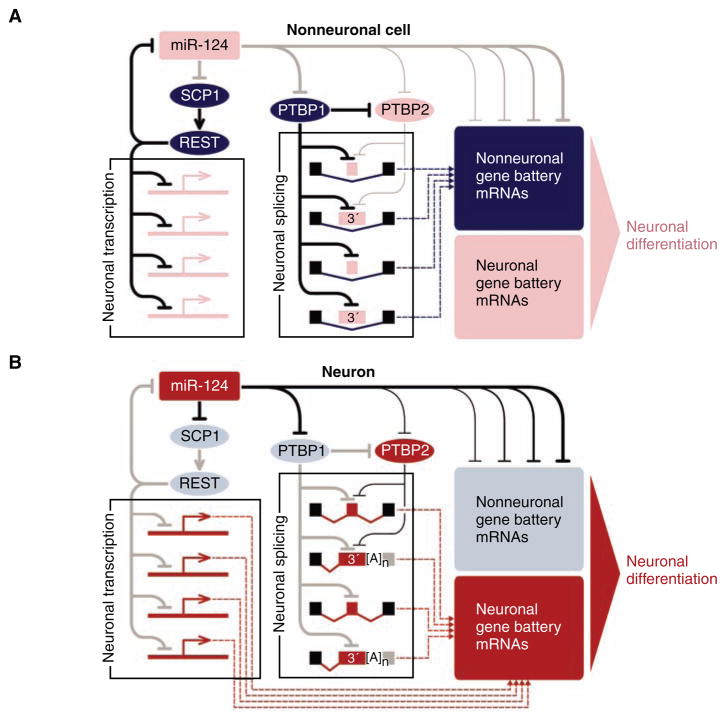
Fig. 1.
MiR-124 controls an extensive gene regulatory network. Active regulatory interactions are shown as solid black lines. Inactive interactions are in gray. Weaker regulatory interactions are rendered as thinner lines. Gene battery expression inputs to the cellular transcriptome are indicated as dashed lines. Nonneuronal elements are colored in dark blue (present) or light blue (absent); neuron-specific elements are colored in corresponding shades of red. This is a simplified diagram that does not account for potential inputs from other neuron-specific miRNAs, as well as the recently identified miRNA function in translational activation (15). Furthermore, PTBP1 and PTBP2 may also activate subsets of alternative exons by using a yet-to-be understood mechanism (19). (A) In nonneuronal cells or neural precursors, miR-124 is either absent or present at a low amount, which allows efficient expression of global repressors of neuronal transcription and alternative pre-mRNA splicing, as well as nonneuronal gene batteries. (B) In differentiating neurons, miR-124 quantities increase, leading to the down-regulation of corresponding repressors, thus allowing the production of the corresponding neuronal proteins. Inversely, the nonneuronal battery genes are directly down-regulated by miR-124.