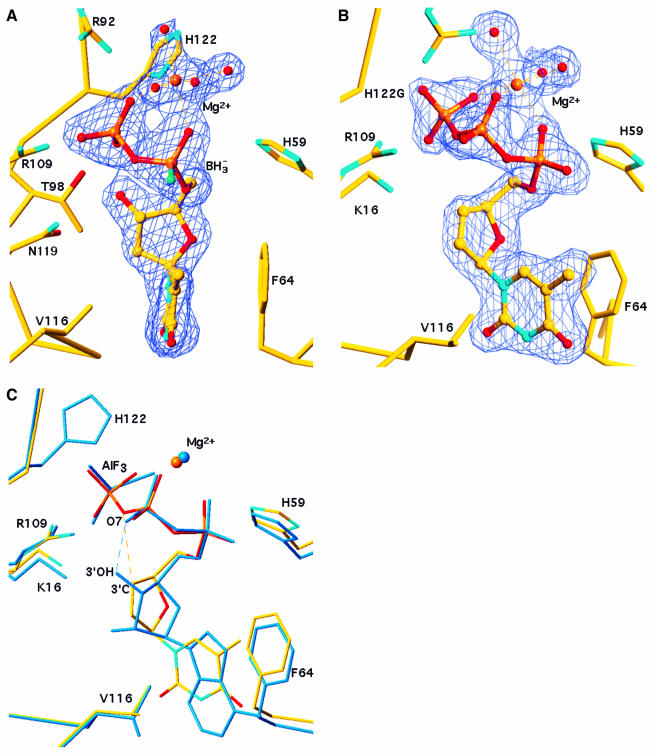
Fig. 2. Nucleotide analogues bound to NDPK. (A) α-(Rp)-borano thymidine diphosphate bound to the wild-type Dictyostelium enzyme. The 2Fo – Fc electron density map at 1.92 Å resolution is contoured at 1σ. The borano group (BH3– in green) points towards the reader. An Mg2+ ion ligates the α- and the β-phosphate, and four water molecules (red spheres) complete the octahedral geometry. The thymine base is sandwiched between F64 and V116. The geometry of the boranophosphate group was taken from the crystal structure of a dimethyl ester (Summers et al., 1998) where the P–B and P–O bond lengths are 1.90 and 1.51 Å, respectively. (B) d4T triphosphate bound to the H122G variant. The 2Fo – Fc electron density map at 1.85 Å resolution is contoured at 1σ. The Mg2+ ion ligates all three phosphates. (C) Comparison with the natural substrate in the ADP–AlF3 complex (PDB file 1KDN; Xu et al., 1997b). Bonds in d4T triphosphate are atom-coloured, the ADP–AlF3 complex is in blue. AlF3 mimics the γ-phosphate undergoing transfer to His122, on top of the figure. In ADP, a hydrogen bond links the 3′-OH of the ribose to oxygen O7 of the β-phosphate. In d4T, the 3′-CH group makes a short (3.2 Å) contact with the equivalent oxygen, which can be interpreted as a CH…O bond (dashes). The superposition is based on all Cα positions. The figure was made with TURBO (Roussel and Cambillau, 1991).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.