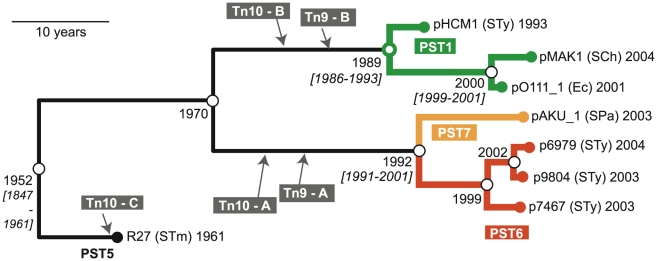
Figure 1. Phylogenetic tree for IncHI1 plasmid sequences.
Phylogenetic tree based on 347 SNPs identified among 8 publicly available IncHI1 plasmid sequences (Table 2), constructed using BEAST (with 20 million iterations, 4 replicate runs, exponential clock model). Terminal nodes are labelled with the organism of origin (STy = Salmonella enterica serovar Typhi, SCh = Salmonella enterica serovar Choleraesuis, STm = Salmonella enterica serovar Typhimurium, SPa = Salmonella enterica serovar Paratyphi A, Ec = E. coli O111:H-) and date of isolation. Isolation dates were input into the BEAST model in order to estimate divergence dates for internal nodes (open circles, labelled with divergence date estimates; brackets indicate 95% highest posterior density interval). Insertion sites (grey) are based on sequence data and verified (except for pO111_1 and pMAK1) by PCR. Precise insertion sites and PCR primers for verification are given in Tables 3 & 4. Four major plasmid groups, PST1, PST5, PST6, PST7, are coloured as labelled.