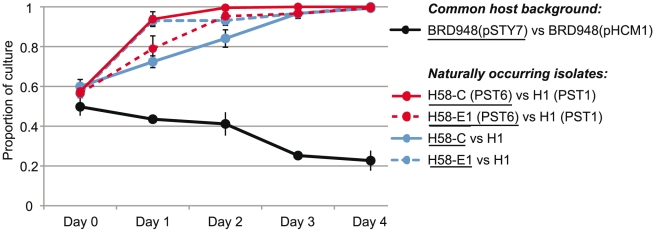
Figure 5. Competitive growth assays for S. Typhi H58 and H1 with and without IncHI1 plasmids.
The dynamics of five competitive growth assays conducted over four days of sequential sub-culture. Black line indicates competition in a common host background (attenuated laboratory strain S. Typhi BRD948; haplotype H10); the proportion of PST1- and PST6-bearing bacteria at each time point was calculated by streaking an aliquot of the sample onto agar plates and testing random colonies using a PCR that differentiates PST1 and PST6. Coloured lines indicate competition between wildtype S. Typhi isolates as specified in the legend (see Table S1 for isolate names); the proportion of H58 and H1 chromosomes at each time point was calculated by quantifying the relative abundance of two alleles at a SNP locus that differs between H58 and H1 S. Typhi using quantitative PCR. For all assays, experiments were replicated at least three times; data points represent the mean proportion of culture corresponding to the isolate underlined in the legend; error bars show the standard deviation of this proportion.