Abstract
Whole genome shotgun sequencing is now possible for extinct organisms, as well as the targeted capture of specific regions. However, targeted resequencing of megabase sized parts of nuclear genomes has yet to be demonstrated for ancient DNA. Here we show that hybridization capture on microarrays can be used to generate large scale targeted data from Neandertal DNA even in the presence of ~99.8% microbial DNA. It is thus now possible to generate high quality data from large regions of the nuclear genome from Neandertals and other extinct organisms. Using this approach we have sequenced ~14,000 protein coding positions that have been inferred to have changed on the human lineage since the last common ancestor shared with chimpanzees. We identify 88 amino acid substitutions that have become fixed in all humans since the divergence from the Neandertals.
The fossil record provides a rough chronological overview of the major phenotypic changes during human evolution. However, the underlying genetic bases for most of these events remain elusive. This is partly because it cannot currently be determined when most human-specific genetic changes, identified from genome comparisons to living relatives, occurred during the ~6.5 million years since the separation of the human and chimpanzee evolutionary lineages. However, recent technological progress has allowed shotgun sequencing to ~1.3-fold coverage of the entire genome of the Neandertal, a human form whose ancestors split from modern human ancestors 200,000 to 330,000 years ago (1). Comparison of Neandertal and present-day human genomes can reveal information about whether genetic changes occurred before or after the ancestral population split of modern humans and Neandertals. However, low coverage whole-genome shotgun sequencing inevitably leaves a substantial proportion of the genome uncovered. While deeper shotgun sequencing of one or a few individuals may produce higher coverage across the whole genome, simple shotgun approaches cannot economically retrieve specific loci from multiple individuals, due to the very high proportion (up to 99.9%) of microbial DNA in most ancient bones. Recently we developed a primer extension capture method to isolate specific DNA sequences from multiple Neandertal individuals (2). However, while useful for capture of small target regions such as mtDNA (2, 3), this method is unlikely to be scalable up to megabase target regions, ruling out experiments such as the retrieval of exomes, large chromosomal regions, or validation of sites of interest identified in the low coverage shotgun genome data.
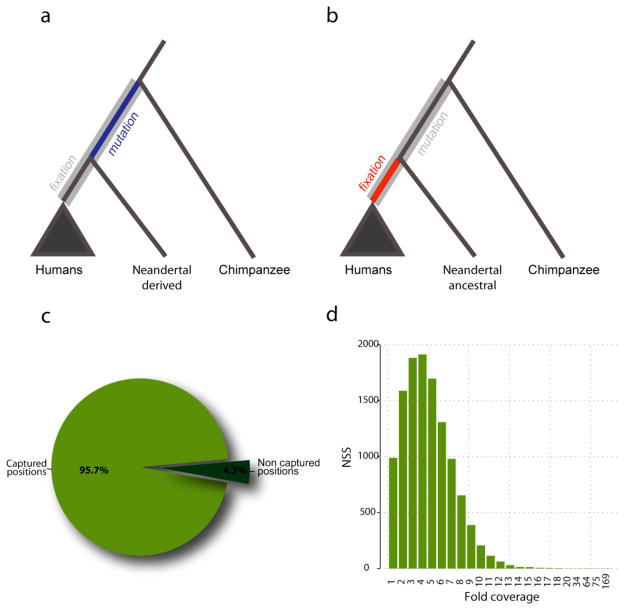
Massively parallel hybridization capture on glass slide microarrays (4, 5) might be a suitable way to capture large regions of the genome from Neandertal individuals, since microarrays can carry hundreds of thousands of probes. Here we demonstrate the power of array capture by isolating Neandertal DNA at thousands of genomic positions where nucleotide substitutions changing amino acids (non-synonymous substitutions) have occurred on the human lineage since its split from chimpanzee. For any such substitution that is fixed, i.e. occur in all present-day humans, it is currently impossible to judge how long ago either the original mutation or the subsequent fixation event occurred. However, by ascertaining the Neandertal state at these positions, we can separate fixed substitutions into, two classes: a) sites where Neandertal carries the derived state; this indicates that the substitution must have occurred before the population split of modern humans and Neandertals ~300,000 years ago (Fig 1a); and b) sites where Neandertal is ancestral; this indicates that fixation of a substitution in modern humans occurred after the population split with Neandertals (Fig 1b).
Figure 1.
(a) For the 9,525 substitutions where the Neandertal carries the derived human allele, the substitution has occurred prior the Neandertal-modern human split (blue part of the hominin lineage), whereas fixation could have happen any time later (grey part of the hominin lineage). (b) For the 88 substitutions where the Neandertal carries the ancestral, chimpanzee-like base, the fixation occurred after the Neandertal-modern human split (red part of the hominin lineage), whereas the substitution could have happened at any point along the hominin lineage (grey). (c) Percentage of the 13,841 positions captured. (d) Distribution of coverage for the positions that encode amino acid substitutions along the hominin lineage. The average coverage is 4.8-fold.
To identify non-synonymous substitutions that occurred on the human lineage since the ancestral split with chimpanzee, we aligned human, chimpanzee, and orangutan protein sequence for all orthologous proteins in HomoloGene (6) [Supporting online material (SOM)]. Comparison of these three species allowed us to assign human/chimpanzee differences to their respective evolutionary lineages. We designed a 1 Million Agilent oligonucleotide array to cover, at 3bp tiling, all the 13,841 substitutions that we inferred to have occurred on the human lineage using the reference human genome. The targeted regions amounted to 1.3Mb (SOM). We used this array to capture target sequences from DNA of a ~49,000-year-old Neandertal bone (Sidron 1253) from El Sidron cave, Spain (7). This bone contains a high amount of Neandertal DNA in absolute terms, but also a high proportion (99.8%) of microbial DNA (2), making it unsuitable for shotgun sequencing. To identify which of the 13,841 substitutions are fixed in present-day humans we also collected data from 50 individuals from the Human Genome Diversity Panel (HGDP) using the same array design used for the Neandertal (Table S1). The DNA libraries from these individuals were barcoded, pooled and captured on a single array. All captured products were sequenced on the Illumina GAII platform and aligned to the human genome (SOM). Overall 37% of the sequence reads aligned to the target regions, representing ~ 190,000-fold target enrichment. We retrieved Neandertal sequence for 13,287 (96%) of the substitutions targeted on the array, with an average coverage of 4.8-fold after filtering for PCR duplicates (Fig. 1c–d). We considered a Neandertal position ancestral if all overlapping reads matched the chimpanzee state and derived if all reads carried the modern human state, disregarding reads that carried a third state or positions where Neandertal reads were found both in the ancestral and in the derived state. From each present-day individual we retrieved on average of 98% (97–99%) of positions with a targeted substitution with an average coverage of 10-fold (Fig. S1). We called genotypes for each individual and considered a position to be fixed derived if it was homozygous and derived in all humans observed, and data was available for at least 25 individuals (50 chromosomes) (SOM).
We included several additional target regions on the array to assess levels of human DNA contamination, which can frequently affect ancient DNA experiments (8). One such region was the complete human mtDNA, which is known to differ between the Sidron 1253 Neandertal analyzed here and present-day humans at 130 positions (2). Despite the fact that the array probes were designed to match present-day human mtDNA, 253,549 of the 254,296 (99.71%) fragments that overlapped these 130 positions matched the Neandertal state. We therefore conclude that the vast majority of mtDNA in the Sidron 1253 library is of Neandertal origin.
For a more direct estimate of contamination in the nuclear DNA we used 46 nucleotide sites on the X chromosome that differ between present-day humans and chimpanzees and that were found to be ancestral in a Neandertal from Croatia (Vindija 33.16) (9) by shotgun sequencing while ~1000 present-day humans in the human diversity panel carry a derived state. The Sidron 1253 individual will obviously not match Vindija 33.16 at all of these sites. However, since Sidron 1253 is a male (10) and thus carried a single X chromosome, at sites where he does match Vindija 33.16, all reads should carry the ancestral base while apparent heterozygosity will indicate human DNA contamination. By analyzing the consistency of reads overlapping these sites on the X chromosome, we calculated a maximum likelihood estimator of X chromosomal contamination of 4%, although confidence intervals are large (1–12%) due to the small number of relevant positions (SOM).
Another way to estimate contamination across autosomes is to investigate patterns of allele counts. Since at every site an individual is either homozygous derived, homozygous ancestral, or heterozygous, DNA from a single individual will yield at each site either only derived alleles, only ancestral alleles or a draw with equal chance for either. Contamination from other individuals would cause systematic deviation from these patterns. Using this idea we produced a likelihood model that estimated contamination at the positions recovered from Sidron 1253, and calculated a 95% upper bound for contamination of 2% (SOM). From these results we conclude that the Sidron 1253 data are not substantially affected by human DNA contamination.
In total, we determined with high confidence the Neandertal and present-day human state for 10,592 non-synonymous substitutions. In 10,015 (91.5%) of all cases the Neandertal carries the derived state, whereas in 937 (8.5%) cases the ancestral state was found (Fig. S2). Of the positions that are fixed in the derived state in present-day humans, 9,525 (87%) are derived in Neandertal while 88 (0.8%) (Table S2) are ancestral (Fig. S2). In agreement with previous results generated by PCR (10), two substitutions that change amino acids in the gene FOXP2, involved in speech and language (11), are both derived in this Neandertal individual.
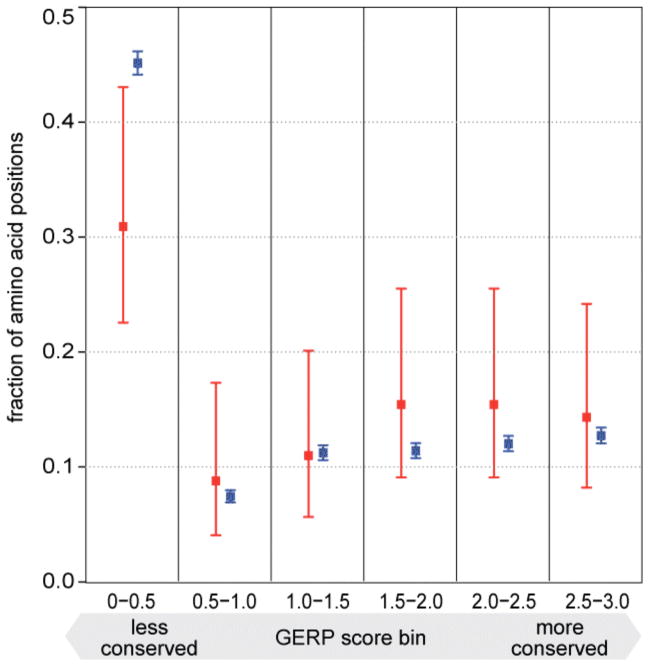
The 88 recently fixed substitutions occur in 83 genes (Table S2–S3). We asked if these genes cluster in any group of functionally related genes (12) (as defined in the Gene Ontology) but found no such groups. We furthermore asked if the 88 substitutions that recently became fixed in humans differ from those where the substitution occurred before the divergence from the Neandertal with respect to how evolutionarily conserved the positions in the encoded proteins are (13) (SOM) (Fig. 2). We find that the 88 recent substitutions tend to affect more amino acid positions that are more conserved than the older substitutions (Wilcoxon rank text; p-value: 0.014). Similarly, the recently fixed substitutions caused more radical amino acid changes with respect to the chemical properties of the amino acids (Wilcoxon rank test; p-value: 0.04). One possible explanation for these observations is that the effective population size of humans since their separation form the Neandertal lineage has been small, leading to a reduced efficiency of purifying selection, as seen e.g. in Europeans (14). We also looked for evidence that the recent substitutions may have been fixed by positive selection. One recent substitution lies in SCML1, a gene involved in spermatogenesis (15) that has been previously proposed as a target of positive selection in humans (16) as well as frequent positive selection in primates (17). However, we found no significant overrepresentation of the 83 genes containing recent substitutions among candidate genes from three genome wide scans for positive selection (18) (Table S4). Nevertheless, we believe that many or all of these amino acid substitutions may warrant functional studies in the future in order to elucidate if they have functional consequences e.g. (19).
Figure 2.
Evolutionary protein conservation at positions affected by the substitutions studied. For each bin of conservation GERP scores, the fractions of amino acid substitutions where the Neandertal carry the derived allele (blue) and the ancestral allele (red) are shown.
Our results demonstrate that hybridization capture arrays can generate data from genomic target regions of megabase size from ancient DNA samples, even when only ~0.2% of the DNA in a sample stems from the endogenous genome. By generating an average coverage of 4- to 5-fold, errors from sequencing and small amounts of human DNA contamination can be minimized, especially in conjunction with the enzymatic removal of uracil residues that are frequent in ancient DNA (20). Since the Sidron 1253 Neandertal library used for this study has been amplified and effectively immortalized, the same library should be able to provide similar quality data for any other genomic target region, or even the entire single-copy fraction of the Neandertal genome.
Supplementary Material
Figures S1. DNA capture of 50 HGDP individuals. (a) Percentage of NSS captured for each of the 50 HGDP individuals. (b) Distribution of coverage for all HGDP individuals.
Figure S2. Classification of NSS using Neandertal sequence. In blue are the fixed NSS where Neandertal has the derived allele (matching the reference human). These substitutions are inferred to have occurred in hominin evolution prior to the Neandertal/human split. As expected, this accounts for the majority of hominin NSS. In red are the fixed NSS where the Neandertal hast the ancestral/chimpanzee base. These fixations are inferred to have occurred since the Neandertal/human split. In light blue and light red are the numbers of NSS where present-day humans within the HGDP panel are still observed to be polymorphic. For the majority of positions where Neandertal is ancestral, present-day humans are polymorphic with the alleles segregating in worldwide populations.
Figure S3. Left: likelihood surface for contamination by heterozygosity at human fixed derived sites. Right: constrained likelihood surface, where heterozygosity is kept constant.
Figure S4. Left: likelihood surface base on positions on the X chromosome for contamination by fraction of Neandertal ancestral at human fixed derived sites. Right: constrained likelihood surface, where heterozygosity is kept constant.
Table S1. Human Genome Diversity Cell Line Panel individuals from whom genomic libraries were captured and sequenced.
Table S2. Recently fixed NSS
Table S3. Genes with 2 recently fixed NSS
Table S4. Overlap NSS and genome wide scans for positive selection
Acknowledgments
We thank Claudia S. Burbano, Cesare de Filippo, Janet Kelso, and David Reich for helpful comments; Martin Kircher and Udo Stenzel for expert technical support; Carlos D. Bustamante and Kirk E. Lohmueller for access to human resequencing databases; David L. Goode and Arendt Sidow for providing conservation scores; Joshua M. Akey for providing coordinates of genome wide scans for selection; Ivo Gut for human genotyping; Emily Leproust and Maethreyan Srinivasan for providing early access to the 1 Million feature Agilent microarrays. The government of the Principado de Asturias funded excavations at the Sidron site. JMG was supported during this research by an NSF international postdoctoral fellowship (OISE-0754461). EH is supported by a postdoctoral training grant from the NIH and by a gift from the Stanley foundation. GJH is an investigator of the Howard Hughes Medical Institute which together with the Presidential Innovation Fund of the Max Planck Society provided generous financial support.
References
- 1.Green RE, et al. 2010 Submited Manuscript. [Google Scholar]
- 2.Briggs AW, et al. Science. 2009 Jul 17;325:318. doi: 10.1126/science.1174462. [DOI] [PubMed] [Google Scholar]
- 3.Krause J, et al. Curr Biol. 2009 Dec 30; [Google Scholar]
- 4.Hodges E, et al. Nat Protoc. 2009;4:960. doi: 10.1038/nprot.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodges E, et al. Nat Genet. 2007 Dec;39:1522. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 6.Sayers EW, et al. Nucleic Acids Res. 2010 Jan;38:D5. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosas A, et al. Proc Natl Acad Sci U S A. 2006 Dec 19;103:19266. doi: 10.1073/pnas.0609662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green RE, et al. EMBO J. 2009 Sep 2;28:2494. doi: 10.1038/emboj.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malez M, Ullrich H. Palaeontologia Jugoslavia. 1982;29:1. [Google Scholar]
- 10.Krause J, et al. Curr Biol. 2007 Nov 6;17:1908. doi: 10.1016/j.cub.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Vargha-Khadem F, Gadian DG, Copp A, Mishkin M. Nat Rev Neurosci. 2005 Feb;6:131. doi: 10.1038/nrn1605. [DOI] [PubMed] [Google Scholar]
- 12.Prufer K, et al. BMC Bioinformatics. 2007;8:41. doi: 10.1186/1471-2105-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper GM, et al. Genome Res. 2005 Jul;15:901. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lohmueller KE, et al. Nature. 2008 Feb 21;451:994. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boeckmann B, et al. Nucleic Acids Res. 2003 Jan 1;31:365. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bustamante CD, et al. Nature. 2005 Oct 20;437:1153. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 17.Wu HH, Su B. BMC Evol Biol. 2008;8:192. doi: 10.1186/1471-2148-8-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akey JM. Genome Res. 2009 May;19:711. doi: 10.1101/gr.086652.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gralle M, Paabo S. 2010 Submited Manuscript. [Google Scholar]
- 20.Briggs AW, et al. Nucleic Acids Res. 2009 Dec 22; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1. DNA capture of 50 HGDP individuals. (a) Percentage of NSS captured for each of the 50 HGDP individuals. (b) Distribution of coverage for all HGDP individuals.
Figure S2. Classification of NSS using Neandertal sequence. In blue are the fixed NSS where Neandertal has the derived allele (matching the reference human). These substitutions are inferred to have occurred in hominin evolution prior to the Neandertal/human split. As expected, this accounts for the majority of hominin NSS. In red are the fixed NSS where the Neandertal hast the ancestral/chimpanzee base. These fixations are inferred to have occurred since the Neandertal/human split. In light blue and light red are the numbers of NSS where present-day humans within the HGDP panel are still observed to be polymorphic. For the majority of positions where Neandertal is ancestral, present-day humans are polymorphic with the alleles segregating in worldwide populations.
Figure S3. Left: likelihood surface for contamination by heterozygosity at human fixed derived sites. Right: constrained likelihood surface, where heterozygosity is kept constant.
Figure S4. Left: likelihood surface base on positions on the X chromosome for contamination by fraction of Neandertal ancestral at human fixed derived sites. Right: constrained likelihood surface, where heterozygosity is kept constant.
Table S1. Human Genome Diversity Cell Line Panel individuals from whom genomic libraries were captured and sequenced.
Table S2. Recently fixed NSS
Table S3. Genes with 2 recently fixed NSS
Table S4. Overlap NSS and genome wide scans for positive selection